More than 300 people across academia and industry spilled into an auditorium to attend a BoltzGen seminar on Thursday, Oct. 30, hosted by the Abdul Latif Jameel Clinic for Machine Learning in Health (MIT Jameel Clinic). Headlining the event was MIT PhD student and BoltzGen’s first author Hannes Stärk, who had announced BoltzGen just a few days prior.
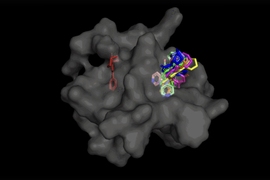
Building upon Boltz-2, an open-source biomolecular structure prediction model predicting protein binding affinity that made waves over the summer, BoltzGen (officially released on Sunday, Oct. 26.) is the first model of its kind to go a step further by generating novel protein binders that are ready to enter the drug discovery pipeline.
Three key innovations make this possible: first, BoltzGen’s ability to carry out a variety of tasks, unifying protein design and structure prediction while maintaining state-of-the-art performance. Next, BoltzGen’s built-in constraints are designed with feedback from wetlab collaborators to ensure the model creates functional proteins that don’t defy the laws of physics or chemistry. Lastly, a rigorous evaluation process tests the model on “undruggable” disease targets, pushing the limits of BoltzGen’s binder generation capabilities.
Most models used in industry or academia are capable of either structure prediction or protein design. Moreover, they’re limited to generating certain types of proteins that bind successfully to easy “targets.” Much like students responding to a test question that looks like their homework, as long as the training data looks similar to the target during binder design, the models often work. But existing methods are nearly always evaluated on targets for which structures with binders already exist, and end up faltering in performance when used on more challenging targets.
“There have been models trying to tackle binder design, but the problem is that these models are modality-specific,” Stärk points out. “A general model does not only mean that we can address more tasks. Additionally, we obtain a better model for the individual task since emulating physics is learned by example, and with a more general training scheme, we provide more such examples containing generalizable physical patterns.”
The BoltzGen researchers went out of their way to test BoltzGen on 26 targets, ranging from therapeutically relevant cases to ones explicitly chosen for their dissimilarity to the training data.
This comprehensive validation process, which took place in eight wetlabs across academia and industry, demonstrates the model’s breadth and potential for breakthrough drug development.
Parabilis Medicines, one of the industry collaborators that tested BoltzGen in a wetlab setting, praised BoltzGen’s potential: “we feel that adopting BoltzGen into our existing Helicon peptide computational platform capabilities promises to accelerate our progress to deliver transformational drugs against major human diseases.”
While the open-source releases of Boltz-1, Boltz-2, and now BoltzGen (which was previewed at the 7th Molecular Machine Learning Conference on Oct. 22) bring new opportunities and transparency in drug development, they also signal that biotech and pharmaceutical industries may need to reevaluate their offerings.
Amid the buzz for BoltzGen on the social media platform X, Justin Grace, a principal machine learning scientist at LabGenius, raised a question. “The private-to-open performance time lag for chat AI systems is [seven] months and falling,” Grace wrote in a post. “It looks to be even shorter in the protein space. How will binder-as-a-service co’s be able to [recoup] investment when we can just wait a few months for the free version?”
For those in academia, BoltzGen represents an expansion and acceleration of scientific possibility. “A question that my students often ask me is, ‘where can AI change the therapeutics game?’” says senior co-author and MIT Professor Regina Barzilay, AI faculty lead for the Jameel Clinic and an affiliate of the Computer Science and Artificial Intelligence Laboratory (CSAIL). “Unless we identify undruggable targets and propose a solution, we won’t be changing the game,” she adds. “The emphasis here is on unsolved problems, which distinguishes Hannes’ work from others in the field.”
Senior co-author Tommi Jaakkola, the Thomas Siebel Professor of Electrical Engineering and Computer Science who is affiliated with the Jameel Clinic and CSAIL, notes that "models such as BoltzGen that are released fully open-source enable broader community-wide efforts to accelerate drug design capabilities.”
Looking ahead, Stärk believes that the future of biomolecular design will be upended by AI models. “I want to build tools that help us manipulate biology to solve disease, or perform tasks with molecular machines that we have not even imagined yet,” he says. “I want to provide these tools and enable biologists to imagine things that they have not even thought of before.”