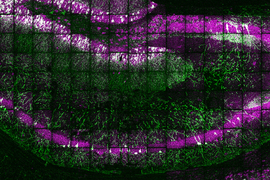
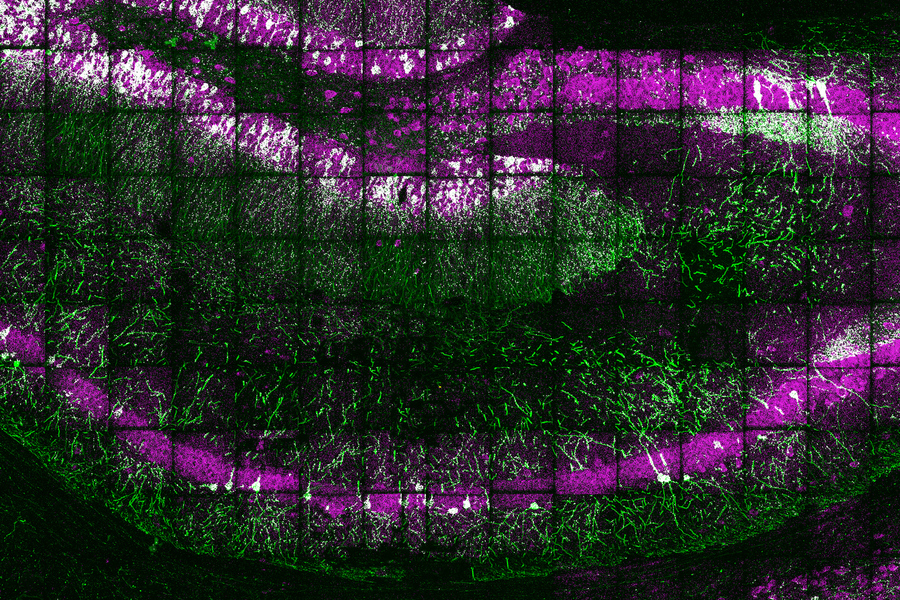
Using a novel technique for expanding tissue, MIT and Harvard Medical School researchers have devised a way to label individual molecules of messenger RNA within a tissue sample and then sequence the RNA.
This approach offers a unique snapshot of which genes are being expressed in different parts of a cell, and could allow scientists to learn much more about how gene expression is influenced by a cell’s location or its interactions with nearby cells. The technique could also be useful for mapping cells in the brain or other tissues and classifying them according to their function.
“Gene expression is one of the most fundamental processes in all of biology, and it plays roles in all biological processes, both healthy and disease-related. However, you need to know more than just whether a gene is on or off,” says Ed Boyden, the Y. Eva Tan Professor in Neurotechnology and a professor of biological engineering, media arts and sciences, and brain and cognitive sciences at MIT. “You want to know where the gene products are located. You care what cell types they’re in, which individual cells they play roles in, and even which parts of cells they work in.”
In a study appearing today in Science, the researchers showed that they could use this technique to locate and then sequence thousands of different messenger RNA molecules within the mouse brain and in human tumor samples.
The senior authors of the study are Boyden, an investigator at the MIT McGovern Institute and the Howard Hughes Medical Institute; George Church, a professor of genetics at Harvard Medical School; and Adam Marblestone, a former MIT research scientist. The paper’s lead authors are Shahar Alon, a former MIT postdoc who is now a senior lecturer at Bar-Ilan University; Daniel Goodwin, an MIT graduate student; Anubhav Sinha ’14 MNG ’15, an MIT graduate student; Asmamaw Wassie ’12, PhD ’19; and Fei Chen PhD ’17, who is an assistant professor of stem cell and regenerative biology at Harvard University and a member of the Broad Institute of MIT and Harvard.
Tissue expansion
The new sequencing technique builds on a method that Boyden’s group devised in 2015 for expanding tissue samples and then imaging them. By embedding water-absorbent polymers into a tissue sample, researchers can swell the tissue sample while keeping its overall organization intact. Using this approach, tissues can be expanded by a factor of 100 or more, allowing scientists to obtain very high-resolution images of the brain or other tissues using a regular light microscope.
In 2014, Church’s lab developed an RNA sequencing technique known as FISSEQ (fluorescent in situ sequencing), which allows thousands of mRNA molecules to be located and sequenced within cells grown in a lab dish. The Boyden and Church labs decided to join forces to combine tissue expansion and in situ RNA sequencing, creating a new technique they call expansion sequencing (ExSeq).
Expanding the tissue before performing RNA sequencing has two main benefits: It offers a higher-resolution look at the RNA in cells, and it makes it easier to sequence those RNA molecules. “When you separate these molecules in the expanding sample, and move them away from each other, that gives you more room to actually perform the chemical reactions of in situ sequencing,” Marblestone says.
Once the tissue is expanded, the researchers can label and sequence thousands of RNA molecules in a sample, at a resolution that allows them to pinpoint the molecules’ locations not only within cells but within specific compartments such as dendrites — the tiny extensions of neurons that receive communications from other neurons.
“We know that the location of RNA in these small regions is important for learning and memory, but until now, we didn't have any way to measure these locations because they are very small, on the order of nanometers,” Alon says.
Using an “untargeted” version of this technique, meaning that they are not looking for specific RNA sequences, the researchers can turn up thousands of different sequences. They estimate that in a given sample, they can sequence between 20 and 50 percent of all of the genes present.
In the mouse hippocampus, this technique yielded some surprising results. For one, the researchers found mRNA containing introns, which are sections of RNA that are normally edited out of mRNA in the nucleus, in dendrites. They also discovered mRNA molecules encoding transcription factors in the dendrites, which may help with novel forms of dendrite-to-nucleus communication.
“These are just examples of things that we never would have gone looking for intentionally, but now that we can sequence RNA exactly where it is in the neuron, we're able to explore a lot more biology,” Goodwin says.
Cellular interactions
The researchers also showed that they could explore gene expression in a more targeted way, looking for a specific set of RNA sequences that correspond to genes of interest. In the visual cortex of the mouse, the researchers used this approach to classify neurons into different types based on an analysis of 42 different genes that they express.
This technology could also be useful to analyze many other kinds of tissues, such as tumor biopsies. In this paper, the researchers studied breast cancer metastases, which contain many different cell types, including cancer cells and immune cells. The study revealed that these cell types can behave differently depending on their location within a tumor. For example, the researchers found that B cells that were near tumor cells expressed certain inflammatory genes at a higher level than B cells that were farther from tumor cells.
“The tumor microenvironment has been studied in many different contexts for a long time, but it’s been difficult to study it with any depth,” Sinha says. “A cancer biologist can give you a list of 20 or 30 marker genes that will identify most of the cell types in the tissue. Here, since we interrogated 297 different RNA transcripts in the sample, we can ask and answer more detailed questions about gene expression.”
The researchers now plan to further study the interactions between cancer cells and immune cells, as well as gene expression in the brain in healthy and disease states. They also plan to extend their techniques to allow them to map additional types of biomolecules, such as proteins, alongside RNA.
The research was funded, in part, by the National Institutes of Health and the National Science Foundation, as well as by Lisa Yang, John Doerr, the Open Philanthropy Project, Cancer Research UK, the Chan Zuckerberg Initiative Human Cell Atlas pilot program, and HHMI.