Beginning with the invention of the first microscope in the late 1500s, scientists have been trying to peer into preserved cells and tissues with ever-greater magnification. The latest generation of so-called “super-resolution” microscopes can see inside cells with resolution better than 250 nanometers.
A team of researchers from MIT has now taken a novel approach to gaining such high-resolution images: Instead of making their microscopes more powerful, they have discovered a method that enlarges tissue samples by embedding them in a polymer that swells when water is added. This allows specimens to be physically magnified, and then imaged at a much higher resolution.
This technique, which uses inexpensive, commercially available chemicals and microscopes commonly found in research labs, should give many more scientists access to super-resolution imaging, the researchers say.
“Instead of acquiring a new microscope to take images with nanoscale resolution, you can take the images on a regular microscope. You physically make the sample bigger, rather than trying to magnify the rays of light that are emitted by the sample,” says Ed Boyden, an associate professor of biological engineering and brain and cognitive sciences at MIT.
Boyden is the senior author of a paper describing the new method in the Jan. 15 online edition of Science. Lead authors of the paper are graduate students Fei Chen and Paul Tillberg.
Physical magnification
Most microscopes work by using lenses to focus light emitted from a sample into a magnified image. However, this approach has a fundamental limit known as the diffraction limit, which means that it can’t be used to visualize objects much smaller than the wavelength of the light being used. For example, if you are using blue-green light with a wavelength of 500 nanometers, you can’t see anything smaller than 250 nanometers.
“Unfortunately, in biology that’s right where things get interesting,” says Boyden, who is a member of MIT’s Media Lab and McGovern Institute for Brain Research. Protein complexes, molecules that transport payloads in and out of cells, and other cellular activities are all organized at the nanoscale.
Scientists have come up with some “really clever tricks” to overcome this limitation, Boyden says. However, these super-resolution techniques work best with small, thin samples, and take a long time to image large samples. “If you want to map the brain, or understand how cancer cells are organized in a metastasizing tumor, or how immune cells are configured in an autoimmune attack, you have to look at a large piece of tissue with nanoscale precision,” he says.
To achieve this, the MIT team focused its attention on the sample rather than the microscope. Their idea was to make specimens easier to image at high resolution by embedding them in an expandable polymer gel made of polyacrylate, a very absorbent material commonly found in diapers.
Before enlarging the tissue, the researchers first label the cell components or proteins that they want to examine, using an antibody that binds to the chosen targets. This antibody is linked to a fluorescent dye, as well as a chemical anchor that can attach the dye to the polyacrylate chain.
Once the tissue is labeled, the researchers add the precursor to the polyacrylate gel and heat it to form the gel. They then digest the proteins that hold the specimen together, allowing it to expand uniformly. The specimen is then washed in salt-free water to induce a 100-fold expansion in volume. Even though the proteins have been broken apart, the original location of each fluorescent label stays the same relative to the overall structure of the tissue because it is anchored to the polyacrylate gel.
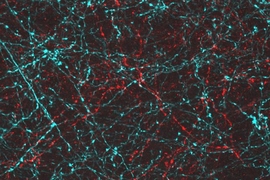
“What you’re left with is a three-dimensional, fluorescent cast of the original material. And the cast itself is swollen, unimpeded by the original biological structure,” Tillberg says.
The MIT team imaged this “cast” with commercially available confocal microscopes, commonly used for fluorescent imaging but usually limited to a resolution of hundreds of nanometers. With their enlarged samples, the researchers achieved resolution down to 70 nanometers. “The expansion microscopy process … should be compatible with many existing microscope designs and systems already in laboratories,” Chen adds.
Large tissue samples
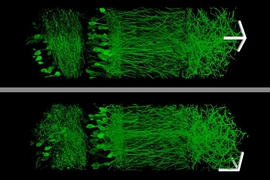
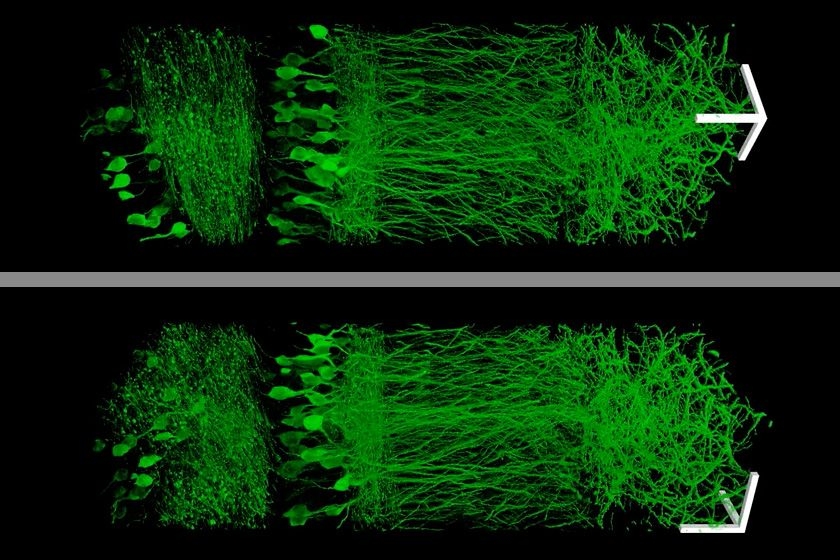
Using this technique, the MIT team was able to image a section of brain tissue 500 by 200 by 100 microns with a standard confocal microscope. Imaging such large samples would not be feasible with other super-resolution techniques, which require minutes to image a tissue slice only 1 micron thick and are limited in their ability to image large samples by optical scattering and other aberrations.
“The exciting part is that this approach can acquire data at the same high speed per pixel as conventional microscopy, contrary to most other methods that beat the diffraction limit for microscopy, which can be 1,000 times slower per pixel,” says George Church, a professor of genetics at Harvard Medical School who was not part of the research team.
“The other methods currently have better resolution, but are harder to use, or slower,” Tillberg says. “The benefits of our method are the ease of use and, more importantly, compatibility with large volumes, which is challenging with existing technologies.”
The researchers envision that this technology could be very useful to scientists trying to image brain cells and map how they connect to each other across large regions.
“There are lots of biological questions where you have to understand a large structure,” Boyden says. “Especially for the brain, you have to be able to image a large volume of tissue, but also to see where all the nanoscale components are.”
While Boyden’s team is focused on the brain, other possible applications for this technique include studying tumor metastasis and angiogenesis (growth of blood vessels to nourish a tumor), or visualizing how immune cells attack specific organs during autoimmune disease.
The research was funded by the National Institutes of Health, the New York Stem Cell Foundation, Jeremy and Joyce Wertheimer, the National Science Foundation, and the Fannie and John Hertz Foundation.