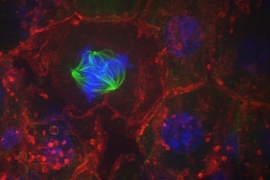
Cells in the body proliferate at different rates. Some divide constantly and throughout life, like the ones that line the gut. Others divide only rarely, sometimes resting for several years in a non-dividing state. Now, a study led by scientists at MIT's Whitehead Institute sheds light on the molecular mechanisms that help control this cellular hibernation, termed quiescence, revealing how cells can purposefully choose to retain the capacity to divide. The team’s findings, which appeared online Aug. 15 in the journal Developmental Cell, hold significance for understanding not just cell division and cell state, but also the dynamics of the cellular machinery that supports these processes, including a group of proteins at a critical structure called the centromere that ensure that chromosomes are properly inherited every time a cell divides.
During cell division, each resident chromosome gets duplicated and then equally apportioned, ensuring that both cells receive a complete set of genetic instructions. The unsung hero of this careful choreography is the centromere, a small chromosomal region that anchors the rope-like fibers that separate chromosomes during cell division. Chromosomes that lack a centromere cannot be transported to their rightful places. That leaves cells with a jumbled mess of DNA — a steppingstone toward disordered growth and, potentially, cancer.
“Our study offers a new perspective on cell identity and cell state,” says senior author Iain Cheeseman, Whitehead Institute member and a professor of biology at MIT. “The key centromere protein, named CENP-A, was widely thought to be static, but is in fact replenished at a slow, yet continuous, rate. This serves not only to refresh and maintain the apparatus required for cell division but also to provide a marker of the cells’ future capacity for proliferation.”
In most organisms, centromeres are not defined by DNA sequence but instead by the assortment of proteins that gather upon them. That is to say, centromeres are spelled out in epigenetic terms. And within the epigenetic lexicon of the centromere, a protein called CENP-A is particularly indispensable. If it is lost, centromeres can never regain the protein and they will malfunction. For that reason, it has been widely believed that CENP-A acts like a boulder — once it lands on the centromere, it never leaves.
“Once you accept the fact that centromeres are demarcated by proteins, you start to imagine the full diversity of situations in which those proteins must remain biologically intact,” says Cheeseman. “And there are some really mind-blowing ones — like human oocytes, which must maintain their centromeres for decades. How does that happen?”
Oocytes, the female reproductive cells, first form in humans during embryonic development and remain dormant until after puberty — representing a decade or more of inactivity. So, does that mean CENP-A just sits there, hanging out on the centromeres, for all those years? That would be a tall order because proteins, just like the parts of a car, tend to wear out and need replacement.
First author Zak Swartz, a postdoc in Cheeseman’s lab, set out to answer this question. Instead of analyzing human oocytes, which are challenging to obtain and cultivate, he devised the methods needed to study sea star oocytes. Remarkably, he and his colleagues discovered that CENP-A is gradually but continuously incorporated into the oocytes’ centromeres over a period of several weeks, reflecting a plodding protein swap that serves to change out old CENP-A proteins for new ones. Notably, when this process is blocked, the centromere proteins are lost and the chromosomes fail to properly position themselves later during oocyte development, a telltale sign of centromere dysfunction that can severely disrupt embryonic development.
“By studying sea star oocytes, with their particular experimental strengths, we were able to reveal a fundamental aspect of biology that had been difficult to notice but is clearly occurring in a wide range of organisms,” says Swartz. “Our findings are a testament to the power of basic science and expanding the diversity of organisms studied in the lab.”
Swartz, Cheeseman, and their colleagues observed a similar CENP-A exchange when they examined other types of quiescent cells, including human cells. But when they studied mature muscle cells — a cell type that has lost its capacity to divide and is therefore at the end of its developmental journey — they uncovered a very different scenario. In these cells, the levels of CENP-A at the centromeres are drastically reduced, particularly when compared to the cells’ younger brethren. The researchers hypothesize that this difference reflects distinct needs to maintain their centromeres, which in turn signals disparate capacities for cell division.
“This suggests that CENP-A is an indicator of proliferative potential,” says Cheeseman. “You could look through the trillions of cells in the body, and if it is there, then cells will be able to segregate their chromosomes; if it isn’t, then they’ll never be able to do so.”
In addition to mapping CENP-A dynamics in different cell types, Cheeseman and his team also uncovered molecular evidence that helps explain how new CENP-A is laid down, particularly in cells that are not actively dividing. As an epigenetic mark, the CENP-A protein forms part of a nucleosome, the unit of bobbin-like histone proteins around which DNA is tightly wound, like thread on a spool. That structure poses some logistical challenges when it comes to incorporating new CENP-A.
The Whitehead Institute team discovered that in quiescent cells, CENP-A deposition requires transcription — the process by which DNA is unspooled from its histones and copied into a chemically similar, single-stranded form. The researchers propose that this chemical conversion provides a destabilizing force that helps to dislodge histones bearing old CENP-A, which allows cells to refresh the old CENP-A molecules by carving out space for their newer counterparts.
Taken together, the team’s findings illuminate the centromere as a carefully groomed structure, even in cells that divide infrequently. The new work has broad implications for the understanding of epigenetic inheritance in normal development and disease, and suggests that defects in centromere maintenance could underlie a range of conditions, from infertility to cancer.
This work was supported by The Harold G & Leila Y. Mathers Charitable Foundation, the NIH/National Institute of General Medical Sciences, American Cancer Society, and the Scott Cook and Signe Ostby fund.