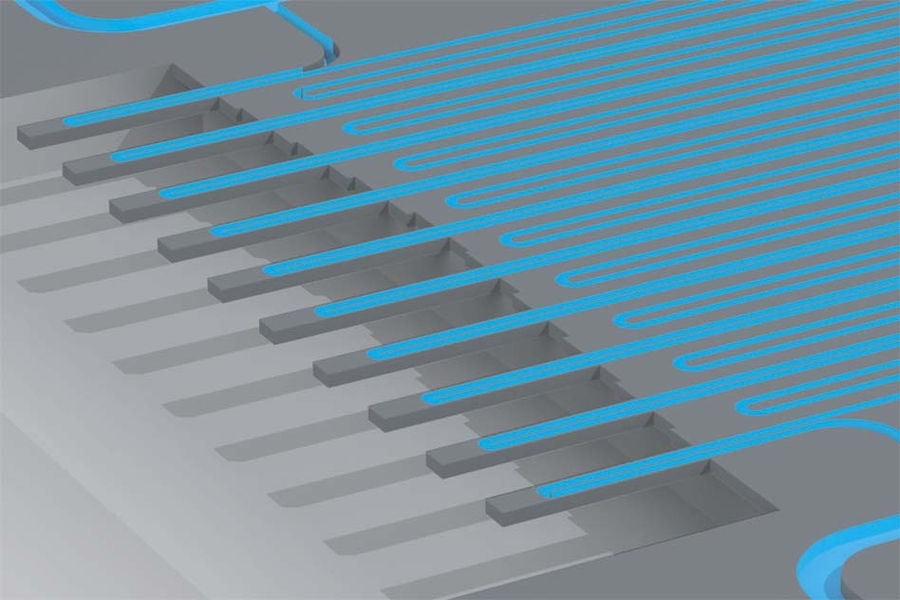
By studying both the physical and genomic features of cancer cells, MIT researchers have come up with a new way to investigate why some cancer cells survive drug treatment while others succumb.
Their new approach, which combines measurements of cell mass and growth rate with analysis of a cell’s gene expression, could be used to reveal new drug targets that would make cancer treatment more effective. Exploiting these targets could help knock out the defenses that cells use to overcome the original drug treatment, the researchers say.
In a paper appearing in the Nov. 28 issue of the journal Genome Biology, the researchers identified a growth signaling pathway that is active in glioblastoma cells that are resistant to an experimental type of drug known as an MDM2 inhibitor.
“By measuring a cell's mass and growth rate immediately prior to single-cell RNA-sequencing, we can now use a cell’s ‘fitness’ to classify it as responsive or nonresponsive to a drug, and to relate this to underlying molecular pathways,” says Alex K. Shalek, the Pfizer-Laubach Career Development Assistant Professor of Chemistry, a member of MIT’s Institute for Medical Engineering and Science (IMES), an extramural member of the Koch Institute for Integrative Cancer Research, and an associate member of the Ragon and Broad Institutes.
Shalek and Scott Manalis, the Andrew and Erna Viterbi Professor in the MIT departments of Biological Engineering and Mechanical Engineering and a member of the Koch Institute, are the senior authors of the study. The paper’s lead author is Robert Kimmerling, a recent MIT PhD recipient.
Cancer cell analysis
About a decade ago, Manalis’ lab invented a technology that allows researchers to measure the mass of single cells. In recent years, they have adapted the device, which measures cells’ masses as they flow through tiny channels, so that it can also measure cell growth rates by repeatedly weighing the cells over short periods of time.
Last year, working with researchers at Dana-Farber Cancer Institute (DFCI), Manalis and his colleagues used this approach to test drug responses of tumor cells from patients with multiple myeloma, a type of blood cancer. After treating the cells with three different drugs, the researchers measured the cells’ growth rates and found they were correlated with the cells’ susceptibility to the treatment.
“Single-cell biophysical properties such as mass and growth rate provide early indicators of drug response, thereby offering the potential to delineate sensitive cells from resistant cells while they are still viable,” Manalis says.
In their new study, the researchers wanted to add a genomic component, which they hoped could help reveal why only certain cells are susceptible to a particular drug. “We wanted to be able to take those measurements and add on some of the biological context for why a cell is growing a certain way or behaving a certain way,” Kimmerling says.
To accomplish this, Kimmerling and Manalis teamed up with Shalek, who has extensive experience in sequencing the messenger RNA (mRNA) of individual cells. This information can provide a snapshot of which genes are being expressed in a single cell at a particular moment.
The researchers modified the cell-weighing system so that cells would be spaced evenly as they flowed through, making it easier to collect them one at a time when they exit the system. The cells are weighed several times over the course of 20 minutes to determine growth rate, and as soon as they reach the end of the channel, they are immediately captured and ruptured to release their RNA for analysis. Shalek’s lab then sequenced the RNA of each of the cells. This approach enabled the mass and growth rate of each cell to be directly linked to its gene expression.
Once they had the system working, the researchers collaborated with Keith Ligon and his lab at DFCI to analyze cancer cells derived from a patient with glioblastoma, an aggressive type of brain cancer. The researchers treated the cells with an MDM2 inhibitor, a type of drug that helps to boost the function of p53, a protein that helps cells stop tumor formation. Such drugs are now in clinical trials to treat glioblastoma. In animal studies, this drug has been effective against tumors, but the tumors often grow back later.
In this study, the researchers hoped to find out why some glioblastoma cells survive MDM2 treatment. They treated the cells, measured their growth rates about 16 hours after the treatment, and then sequenced their RNA. “Before the cells have lost viability, we can measure their mass and their growth rate to reveal drug response heterogeneity to that treatment, and then link that with their gene expression,” Kimmerling says.
Importantly, the researchers found subpopulations of cells that were not responsive to the drug. RNA sequencing revealed that in cells that were responsive, genes required for programmed cell death were turned on. Meanwhile, in cells that did not seem to be vulnerable to the drug, genes involved in mTOR, a signaling pathway involved in growth and survival, were turned up.
“What we’re excited about here is we now have this list of biological targets to look into,” Kimmerling says. “We can start to generate testable hypotheses from these gene expression signatures that are more highly expressed in the cells that continue to grow after drug treatment.”
Possible drug targets
The researchers now plan to explore the possibility of targeting some of the genes that were turned up on the nonresponding cells, in hopes of developing drugs that could be used together with the original MDM2 inhibitor. They also hope to adapt this approach for other types of cancers. Some, such as blood cancers, are easier to study than solid tumors, which are more difficult to separate into single cells.
“The hope is that we’ll be able to apply this technology to any sample that can be dissociated into a single-cell population,” Kimmerling says.
Another possible application of the cell-growth measurement technology is studying tumor cells from individual patients to try to predict how they will respond to a particular drug. Kimmerling, Manalis, and others have founded a company called Travera, which has licensed the technology and hopes to develop it for patient use. The company is currently not working on the RNA sequencing aspect of the technology, but that element could also be valuable to incorporate in the future, Kimmerling says.
The research was funded by the Cancer Systems Biology Consortium U54 Research Center and the Cancer Center Support (core) Grant from the National Cancer Institute; the Searle Scholars Program; the Beckman Young Investigator Program; the National Institutes of Health, including an NIH New Innovator Award; the Pew-Stewart Scholars; and a Sloan Fellowship in Chemistry.