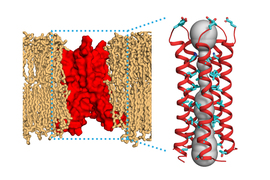
The genome of the SARS-CoV-2 virus encodes 29 proteins, one of which is an ion channel called E. This channel, which transports protons and calcium ions, induces infected cells to launch an inflammatory response that damages tissues and contributes to the symptoms of Covid-19.
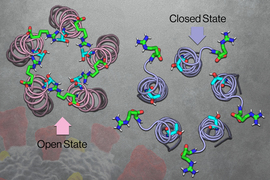
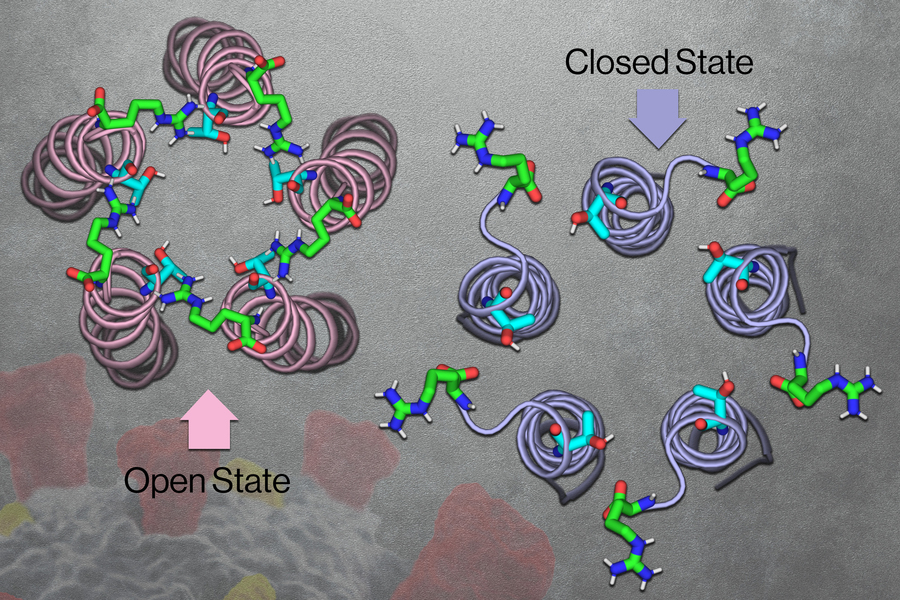
MIT chemists have now discovered the structure of the “open” state of this channel, which allows ions to flow through. This structure, combined with the “closed” state structure that was reported by the same lab in 2020, could help scientists figure out what triggers the channel to open and close. These structures could also guide researchers in developing antiviral drugs that block the channel and help prevent inflammation.
“The E channel is an antiviral drug target. If you can stop the channel from sending calcium into the cytoplasm, then you have a way to reduce the cytotoxic effects of the virus,” says Mei Hong, an MIT professor of chemistry and the senior author of the study.
MIT postdoc Joao Medeiros-Silva is the lead author of the study, which appears today in Science Advances. MIT postdocs Aurelio Dregni and Pu Duan and graduate student Noah Somberg are also authors of the paper.
Open and closed
Hong has extensive experience in studying the structures of proteins that are embedded in cell membranes, so when the Covid-19 pandemic began in 2020, she turned her attention to the coronavirus E channel.
When SARS-CoV-2 infects cells, the E channel embeds itself inside the membrane that surrounds a cellular organelle called the ER-Golgi intermediate compartment (ERGIC). The ERGIC interior has a high concentration of protons and calcium ions, which the E channel transports out of ERGIC and into the cell cytoplasm. That influx of protons and calcium leads to the formation of multiprotein complexes called inflammasomes, which induce inflammation.
To study membrane-embedded proteins such as ion channels, Hong has developed techniques that use nuclear magnetic resonance (NMR) spectroscopy to reveal the atomic-level structures of those proteins. In previous work, her lab used these techniques to discover the structure of an influenza protein known as the M2 proton channel, which, like the coronavirus E protein, consists of a bundle of several helical proteins.
Early in the pandemic, Hong’s lab used NMR to analyze the structure of the coronavirus E channel at neutral pH. The resulting structure, reported in 2020, consisted of five helices tightly bundled together in what appeared to be the closed state of the channel.
“By 2020, we had matured all the NMR technologies to solve the structure of this kind of alpha-helical bundles in the membrane, so we were able to solve the closed E structure in about six months,” Hong says.
Once they established the closed structure, the researchers set out to determine the structure of the open state of the channel. To induce the channel to take the open conformation, the researchers exposed it to a more acidic environment, along with higher calcium ion levels. They found that under these conditions, the top opening of the channel (the part that would extend into the ERGIC) became wider and coated with water molecules. That coating of water makes the channel more inviting for ions to enter.
That pore opening also contains amino acids with hydrophilic side chains that dangle from the channel and help to attract positively charged ions.
The researchers also found that while the closed channel has a very narrow opening at the top and a broader opening at the bottom, the open state is the opposite: broader at the top and narrower at the bottom. The opening at the bottom also contains hydrophilic amino acids that help draw ions through a narrow “hydrophobic gate” in the middle of the channel, allowing the ions to eventually exit into the cytoplasm.
Near the hydrophobic gate, the researchers also discovered a tight “belt,” which consists of three copies of phenylalanine, an amino acid with an aromatic side chain. Depending on how these phenylalanines are arranged, the side chains can either extend into the channel to block it or swing open to allow ions to pass through.
“We think the side chain conformation of these three regularly spaced phenylalanine residues plays an important role in regulating the closed and open state,” Hong says.
Viral targeting
Previous research has shown that when SARS-CoV-2 viruses are mutated so that they don’t produce the E channel, the viruses generate much less inflammation and cause less damage to host cells.
Working with collaborators at the University of California at San Francisco, Hong is now developing molecules that could bind to the E channel and prevent ions from traveling through it, in hopes of generating antiviral drugs that would reduce the inflammation produced by SARS-CoV-2.
Her lab is also planning to investigate how mutations in subsequent variants of SARS-CoV-2 might affect the structure and function of the E channel. In the Omicron variant, one of the hydrophilic, or polar, amino acids found in the pore opening is mutated to a hydrophobic amino acid called isoleucine.
“The E variant in Omicron is something we want to study next,” Hong says. “We can make a mutant and see how disruption of that polar network changes the structural and dynamical aspect of this protein.”
The research was funded by the National Institutes of Health and the MIT School of Science Sloan Fund.