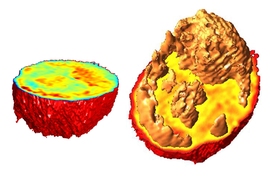
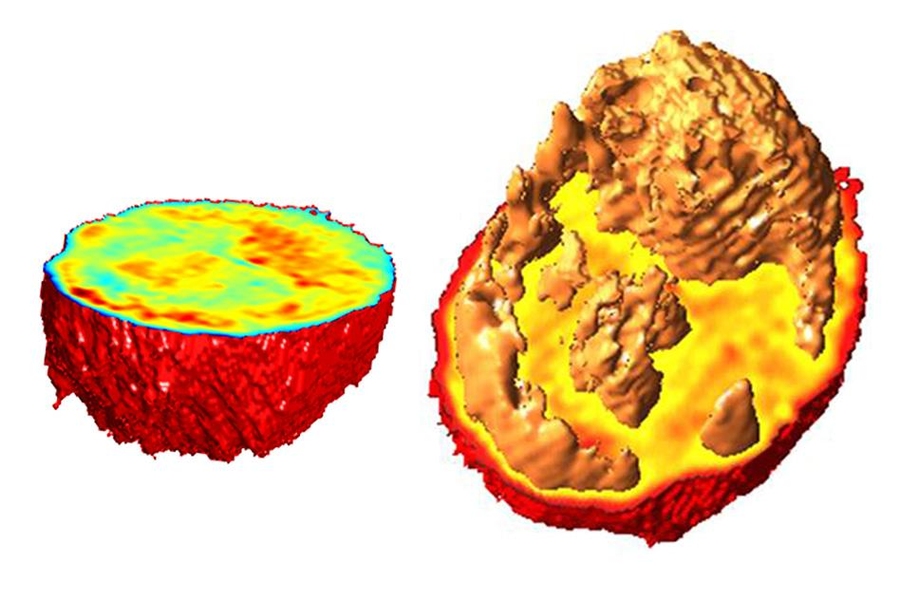
In 2007, MIT scientists developed a type of microscopy that allowed them to detail the interior of a living cell in three dimensions, without adding any fluorescent markers or other labels. This technique also revealed key properties, such as the cells’ density.
Now the researchers have adapted that method so they can image cells as they flow through a tiny microfluidic channel — an important step toward cell-sorting systems that could help scientists separate stem cells at varying stages of development, or to distinguish healthy cells from cancerous cells.
Other sorting methods require scientists to add a fluorescent molecule that highlights the cells of interest, but those tags can damage the cells and make them unsuitable for therapeutic uses.
“Many stem cell applications require sorting of cells at different stages of differentiation. This can be done with fluorescent staining, but once you stain the cells they cannot be used. With our approach, you can utilize a vast amount of information about the 3-D distribution of the cells’ mass to sort them,” says Yongjin Sung, a former postdoc in MIT’s Laser Biomedical Research Center and lead author of a paper describing the technique in the inaugural issue of the journal PRApplied.
Instead of using fluorescent tags, the MIT method analyzes the cells’ index of refraction — a measurement of how much the speed of light is reduced as it passes through a material. Every material has a distinctive index of refraction, and this property can be used, along with cells’ volume, to calculate their mass and density.
Different parts of a cell, including individual organelles, have different indices of refraction, so the information generated by this approach can also be used to identify some of these internal cell structures, such as the nucleus and nucleolus, a structure located within the nucleus.
In the original 2007 version of this technology, known as tomographic phase microscopy, researchers led by the late MIT professor Michael Feld created 3-D images by combining a series of 2-D images taken as laser beams passed through cells from hundreds of different angles. This is the same concept behind CT scanning, which combines X-ray images taken from many different angles to create a 3-D rendering.
For the past few years, Sung and others in the Laser Biomedical Research Center have continued developing the tomographic phase microscopy system. In a paper published in 2012, the MIT team used the technique to measure the mass of chromosomes in living cells; last year, working with Marc Kirschner of Harvard Medical School, the researchers studied volume and density changes in cartilage cells as they differentiated into their final form.
For the new PRApplied study, the MIT researchers collaborated with Daniel Irimia’s lab at Harvard Medical School to adapt the system to image cells as they flow continuously through a microfluidic channel. Other researchers have tried to do this, but their systems required that the cells be halted at a certain point in the channel to complete the imaging. “The only moving part in our system is the sample, which will afford great flexibility in miniaturizing the system,” says Sung, who is now an instructor in radiology at Massachusetts General Hospital.
A key feature of the new MIT system is the use of a focused laser beam that can illuminate cells from many different angles, allowing the researchers to analyze the scattered light from the cells as they flow across the beam. Using a technique known as off-axis digital holography, the researchers can instantaneously record both the amplitude and phase of scattered light at each location of the cells.
“As the cell flows across, we can effectively illuminate the entire sample from all angles without having to rotate a light source or the cell,” says former MIT graduate student Niyom Lue, a coauthor of the new paper.
Changhuei Yang, a professor of electrical engineering and bioengineering at the California Institute of Technology, says the new system is “a very interesting development in the research area of efficient microscopy imaging.”
“The research group very nicely demonstrates that the combination of cell flow as a scanning strategy with line illumination can provide high-quality holographic data for rendering 3-D images of the target cells. As a method, I think this approach has a lot of promise in cytometry type applications,” says Yang, who was not involved in the research.
The current system can image about 10 cells per second, but the researchers hope to speed it up to thousands of cells per second, which would make it useful for applications such as sorting stem cells. The researchers also hope to use the system to learn more about how cancer cells grow and respond to different drug treatments.
“This label-free method can look at different states of the cell, whether they are healthy or whether they maybe have cancer or viral or bacterial infections,” says Peter So, an MIT professor of mechanical engineering and biological engineering who is senior author of the new paper. “We can use this technique to look at the pathological state of the cell, or cells under treatment of some drug, and follow the population over a period of time.”
The research was funded by the National Institutes of Health and Hamamatsu Photonics. Other authors of the paper are MIT principal research scientist Ramachandra Dasari; former MIT postdoc Wonshik Choi; MIT research scientist Zahid Yaqoob; and Bashar Hamza and Joseph Martel of Harvard Medical School.