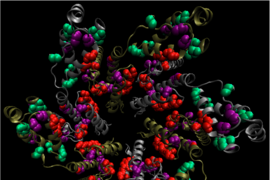
In a new finding that may allow vaccine designers to sidestep part of that obstacle, researchers at the Ragon Institute of Massachusetts General Hospital, MIT and Harvard University have identified sections of an HIV protein where mutations would actually undermine the virus’ fitness — its ability to survive and reproduce.
Vaccines that prime immune cells to specifically target those vulnerable regions could prove much more effective than previously tested vaccines, says Arup Chakraborty, the Robert T. Haslam (1911) Professor at MIT and senior author of a paper on the work appearing in the Proceedings of the National Academy of Sciences the week of June 20.
Though global HIV infection rates have dropped since 2000, there are still more than 33 million people living with AIDS. The vast majority of those people live in developing countries, where there is limited access to antiretroviral drugs that can control the infection.
“Even though we have treatments, the number of people in need globally is outpacing our ability to provide these drugs,” says Harvard Medical School Professor Bruce Walker, director of the Ragon Institute and a senior author of the new paper. “The only real solution is development of an effective vaccine.”
Lead authors of the paper are Vincent Dahirel, a former postdoc in Chakraborty’s lab who is now a professor of chemistry at the Université Marie et Pierre Curie in Paris; and Karthik Shekhar, a chemical engineering PhD student at MIT.
Co-evolution
Viral vaccines usually consist of killed or weakened versions of a virus that prime the body’s immune system to respond when it later encounters the real thing. Most experimental HIV vaccines include some proteins found in the virus’s genetic material.
Vaccines provoke the recipient’s immune system to generate two types of responses: antibodies that can battle viruses in blood or outside cells, and memory T cells, which attack cells that display viral proteins on their surfaces — a sign of infection. However, HIV can escape these responses when its viral proteins evolve to new forms that the vaccine-induced antibodies and T cells no longer recognize. Most researchers believe that an effective HIV vaccine will have to include both an antibody and a T-cell component.
In recent years, designers of the T-cell arm of a vaccine have looked to target single amino acids (the building blocks of proteins) that seem unable to evolve to a different form, with the goal of inducing mutations that incapacitate the virus. So far, this strategy has had limited success, because mutations elsewhere in the viral protein can help restore the loss of fitness.
The Ragon Institute researchers took a broader approach, looking not just at single mutations, but trying to determine if there might be groups of amino acids within viral proteins that evolve together in a coordinated way. After identifying some such groups, they determined whether multiple mutations in those groups tended to be beneficial or harmful to the virus’s survival. A group in which multiple mutations are most harmful could be a good vaccine target, because the virus may undermine its own survival if it tries to mutate those sites, and escape pathways would be limited.
The Ragon team analyzed available HIV protein sequences obtained from infected patients using a mathematical approach, including a method called random matrix theory, which was developed by Eugene Wigner in the 1950s to study high-energy physics. Since then, it has been used in many other areas of physics, but has also been applied in other fields, including economics (to study stock market fluctuations) and biology (to analyze sequences of an enzyme family).
For example, Boston University physicist Eugene Stanley has used random matrix theory to find inherent correlations among the stock prices of companies whose economic activities are coupled. He was able to identify groups of companies whose prices fluctuate collectively, but independently of the fluctuations of other groups of companies. (For example, he found that oil and gas company stock prices fluctuate together, but essentially independently of stock prices in the financial sector.)
Multiple mutations
The Ragon team focused on an HIV polyprotein called Gag, which gives the virus much of its structure, and identified five co-evolving groups of amino acids within the protein. The researchers looked at each pair of sites within the groups, calculating whether a double mutation was beneficial or detrimental to the virus’s survival. (They also analyzed triplets and larger groups.) They discovered that one of the groups, which they term sector 3, had the highest proportion of detrimental multiple mutations.
Structural analysis revealed that amino acids in sector 3 are located at interfaces between proteins that form the viral capsid surrounding the virus’s genetic material. If you make multiple mutations to these amino acids, Chakraborty says, it is difficult for the virus to assemble the capsid.
The Ragon team then tested its findings against human clinical data, discovering that T cells in patients who control HIV without medication do in fact disproportionately target sector-3 amino acids at multiple points, and HIV strains with multiple mutations in this sector are rare, indicating that those strains are less likely to survive.
This finding strengthens the argument that these protein sequences would make good vaccine targets, notes Gregory Petsko, professor of biochemistry and chemistry at Brandeis University. “Tying it to the patient population is what sets this apart, in my mind, from a traditional computational study,” Petsko says.
The Ragon researchers suggest designs for test vaccines based on the vulnerabilities they found in the Gag protein, and are now looking for vulnerable targets in other HIV proteins.
Rafi Ahmed, professor of immunology at the Emory University Vaccine Center, says the paper offers an exciting new approach to designing HIV vaccines. “It breaks new ground in terms of vaccine design and potential insights into why elite controllers are more effective at controlling HIV infection, and it provides additional protein regions to examine,” Ahmed says.