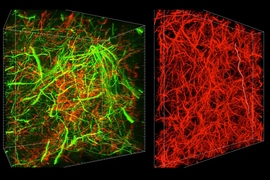
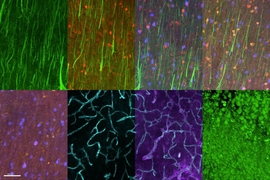
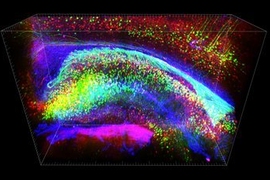
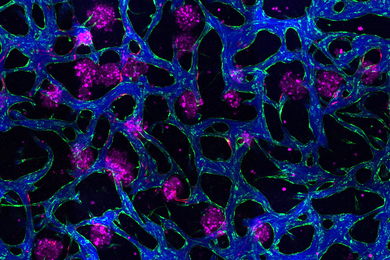
MIT chemical engineers and neuroscientists have devised a new way to preserve biological tissue, allowing them to visualize proteins, DNA, and other molecules within cells, and to map the connections between neurons.
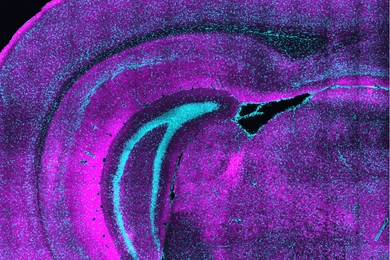
The researchers showed that they could use this method, known as SHIELD, to trace the connections between neurons in a part of the brain that helps control movement and other neurons throughout the brain.
“Using our technique, for the first time, we were able to map the connectivity of these neurons at single-cell resolution,” says Kwanghun Chung, an assistant professor of chemical engineering and a member of MIT’s Institute for Medical Engineering and Science and Picower Institute for Learning and Memory. “We can get all this multiscale, multidimensional information from the same tissue in a fully integrated manner because with SHIELD we can protect all this information.”
Chung is the senior author of the paper, which appears in the Dec. 17 issue of Nature Biotechnology. The paper’s lead authors are MIT postdocs Young-Gyun Park, Chang Ho Sohn, and Ritchie Chen.
Chung is now leading a team of researchers from several institutions that recently received a National Institutes of Health grant to use this technique to produce three-dimensional maps of the entire human brain. “We will be working with the Matthew Frosch group at MGH, the Van Wedeen group at MGH, the Sebastian Seung group at Princeton, and the Laura Brattain group at MIT Lincoln Lab to generate the most comprehensive brain map yet,” he says.
Preserving information
Brain tissue is very delicate and cannot be easily studied unless steps are taken to preserve the tissue from damage. Chung and other researchers have previously developed techniques that allow them to preserve certain molecular components of brain tissue for research, including proteins or messenger RNA, which reveals which genes are turned on.
However, Chung says, “there is no good method that can preserve everything.”
Chung and his colleagues hypothesized that they might be able to better preserve tissue using molecules called polyepoxides — reactive organic molecules that are often used to produce glues. They tested several commercially available polyepoxides and discovered one that had distinctive structural traits that made it ideally suited for their purposes.
The epoxide they chose has a flexible backbone and five branches, each of which can bind to certain amino acids (the building blocks of proteins), as well as other molecules such as DNA and RNA. The flexible backbone allows the epoxides to bind to several spots along the target molecules, and to form cross-links with nearby biomolecules. This renders individual biomolecules and the entire tissue structure very stable and resistant to damage from heat, acid, or other harmful agents. SHIELD also protects key properties of biomolecules, such as protein fluorescence and antigenicity.
To protect large-scale brain tissues and clinical samples, the researchers combined SHIELD with SWITCH, another technique they developed to control chemical reaction speed. They first use the SWITCH-OFF buffer, which halts chemical reactions, to give the epoxides time to diffuse through the entire tissue. When the researchers move the sample to SWITCH-ON condition, the epoxides begin to bind to nearby molecules.
To speed up the clearing and labeling process of SHIELD-protected tissue, the researchers also applied a randomly changing electric field, which they have previously shown increases the transport rate of the molecules. In this paper, they showed that the entire process from preservation to labeling of biopsy tissue could be performed in just four hours.
“We found that this SHIELD coating keeps proteins stable against harsh stressors,” Chung says. “Because we can preserve all the information that we want, and we can extract it at multiple stages, we can better understand the functions of biological components, including neural circuits.”
Once the tissue is preserved, the researchers can label a variety of different targets, including proteins and mRNA produced by the cells. They can also apply techniques such as MAP, which Chung developed in 2016, to expand the tissue and image it at different size scales.
In this paper, the researchers worked with Byungkook Lim’s group at the University of California at San Diego to use SHIELD to map a brain circuit that begins in the globus pallidus externa (GPe), part of the brain’s basal ganglia. This region, which is involved in motor control and other behaviors, is one of the targets of deep brain stimulation — a type of electrical stimulation sometimes used to treat Parkinson’s disease. In the mouse brain, Chung and his colleagues were able to trace the connections between neurons in the GPe and in other parts of the brain, and to count the number of putative synaptic connections between these neurons.
Better biopsies
The speed of SHIELD tissue processing means that it also holds promise for performing rapid, more informative biopsies of patient tissue samples, Chung says. Current methods require embedding tissue samples with paraffin, slicing them, and then applying stains that can reveal cell and tissue abnormalities.
“The current way of doing tissue diagnosis hasn’t changed in many decades, and the process takes days or weeks,” Chung says. “Using our technique, we can rapidly process intact biopsy samples and immuno-label them with really specific, clinically relevant antibodies, and then image the whole thing at high resolution, in three dimensions. And everything can be done in four hours.”
In this paper, the researchers showed that they could label mouse kidney tumor with an antibody that targets proliferating cancer cells.
“The stabilization and preservation of biological information within tissue samples is essential in experiments for optical microscopy,” says Liqun Luo, a professor of biology at Stanford University, who was not involved in the research. “The achievement of SHIELD is not a large advance in just one category, but rather marked improvements across the board, in preserving proteins, transcripts, and tissue structure, as samples are processed through the harsh techniques prescribed by today's best labeling and imaging protocols.”
The MIT team hopes to make this technology widely available and has already distributed it to more than 50 labs around the world. The research was funded by the Burroughs Wellcome Fund Career Award at the Scientific Interface, the Searle Scholars Program, the Packard Award in Science and Engineering, the NARSAD Young Investigator Award, the McKnight Foundation Technology Award, the JPB Foundation, and NCSOFT Cultural Foundation, and the National Institutes of Health.