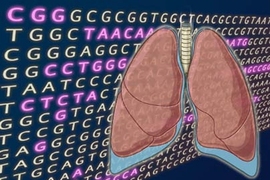
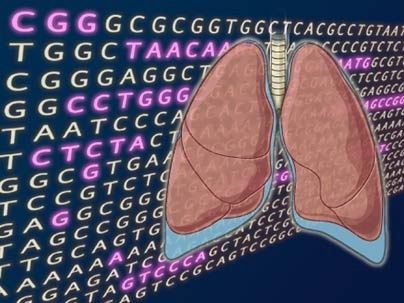
An international team of scientists has produced the most comprehensive view yet of the abnormal genetic landscape of lung cancer, the world's leading cause of cancer deaths. Appearing in the Nov. 4 advance online issue of Nature, the research reveals more than 50 genomic regions that are frequently gained or lost in human lung tumors.
While one-third of these regions contain genes already known to play important roles in lung cancer, the majority harbor new genes yet to be discovered. Flowing from this work, the scientists uncovered a critical gene alteration--not previously linked to any form of cancer--that is implicated in a significant fraction of lung cancer cases, shedding light on the biological basis of the disease and a potential new target for therapy.
"This view of the lung cancer genome is unprecedented, both in its breadth and depth," said senior author Matthew Meyerson, a senior associate member of the Broad Institute of MIT and Harvard and an associate professor at Dana-Farber Cancer Institute and Harvard Medical School. "It lays an essential foundation and has already pinpointed an important gene that controls the growth of lung cells. This information offers crucial inroads to the biology of lung cancer and will help shape new strategies for cancer diagnosis and therapy."
"The genomic landscape of lung cancer gives us a systematic picture of this terrible disease, confirming things we know, but also pointing us to many missing pieces of the puzzle," said Eric Lander, one of the study's co-authors and the founding director of the Broad Institute of MIT and Harvard. "More broadly, the study represents a general approach that can and should be used to analyze all types of cancer. Indeed, the study was designed as a pilot project for an even more comprehensive effort to unearth the genetic causes of cancer."
Each year more than one million people die of lung cancer, including more than 150,000 in the United States. New approaches to treatment rely on a deeper understanding of what goes wrong in cells to spur cancer growth. Through decades of research, it has become clear that lung cancer--like most human cancers--stems mainly from DNA changes that accrue in cells throughout a person's life. But the nature of these changes and their biological consequences remain largely unknown.
To assemble a genome-wide catalog of genetic differences in lung cancer cells, a large-scale project was recently launched in lung adenocarcinoma. The effort, known as the Tumor Sequencing Project (TSP), unites scientists and clinicians throughout the cancer research community.
The TSP researchers studied more than 500 tumor specimens from lung cancer patients. Access to this large collection of high-quality samples made it possible to determine the genetic changes shared among different patients--such recurring changes can highlight important genes involved in cancer growth.
To analyze DNA from each lung tumor, the scientists relied on recent genomic technologies to scan the human genome for hundreds of thousands of genetic markers, called single nucleotide polymorphisms or SNPs. This high-resolution view helped pinpoint which parts of the tumor genome were present in excess copies or missing altogether.
From this work, the researchers uncovered a total of 57 genomic changes that occur frequently in lung cancer patients. Of these, only about 15 are linked to genes previously known to be involved in lung adenocarcinoma. The rest, though, remain to be discovered.
Strikingly, the most common abnormality identified in the study involves a region on chromosome 14 that encompasses two known genes, neither of which had been previously associated with cancer. Through additional studies in cancer cells, co-first author Sue-Ann Woo and other researchers at Dana-Farber Cancer Institute revealed that one of the genes, NKX2.1, influences cancer cell growth. The NKX2.1 gene normally acts as a sort of "master regulator"--controlling the activity of other key genes--in a special group of cells lining the lungs' tiny air sacs, called alveoli. This discovery, that a gene functioning in a select group of cells rather than all cells can promote cancer growth, may have broad implications for the design of novel, molecularly targeted cancer drugs.
The second phase of the TSP, now underway, will examine the same lung tumor samples analyzed in the first phase, but at an even greater level of genetic detail. Using high-throughput DNA sequencing methods, the scientists will characterize small changes in the genetic code of several hundred human genes, which are already implicated in other cancers or more generally in cell growth.
Participating institutions in the TSP include three large-scale DNA sequencing centers--Baylor College of Medicine, Broad Institute of MIT and Harvard, and Washington University--and six medical institutions--Brigham and Women's Hospital, Dana-Farber Cancer Institute, M.D. Anderson Cancer Center, Memorial Sloan-Kettering Cancer Center, the University of Michigan, and Washington University. Investigators from Nagoya City University, the Ontario Cancer Institute/Princess Margaret Hospital, and the University of Texas-Southwestern Medical School also participated in the SNP study.
In addition to Matthew Meyerson and Eric Lander, the scientific leaders of the TSP include Harold Varmus of the Memorial Sloan-Kettering Cancer Center, Richard Gibbs of the Baylor College of Medicine, and Richard Wilson of Washington University in Saint Louis.
The TSP is helping to lay the foundation for future large-scale cancer genome projects, including The Cancer Genome Atlas (TCGA) pilot project. In December 2005, the National Human Genome Research Institute and the National Cancer Institute launched the TCGA pilot to test the feasibility of a comprehensive, systematic approach to exploring the genomics of a wide range of common human cancers.
All data generated by the TSP are being made available to the scientific community in public databases, including caintegrator-info.nci.nih.gov/csp. Data can also be accessed through the Broad Institute web site, at www.broad.mit.edu/tsp.
A version of this article appeared in MIT Tech Talk on November 7, 2007 (download PDF).