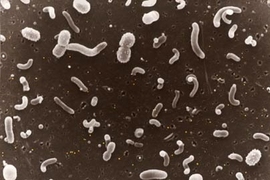
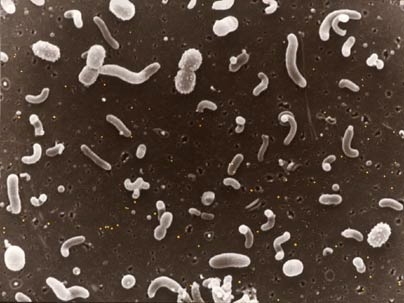
Using DNA analysis, MIT researchers and colleagues have gained new insight into how marine microbes thrive and survive at different depths of the ocean.
"Microbes are the central processors of matter and energy in almost every ecosystem imaginable -- especially so in the sea," said MIT Professor Ed DeLong, who led the work. Thousands of different types of microbes, the world's smallest creatures, inhabit every cubic centimeter of seawater. They have huge effects on ocean chemistry and possibly even climate.
However, "their complex interactions are really tough to study in natural environments," DeLong said. "We took a shortcut to understanding their environmental activities by analyzing the DNA from whole communities of microbes."
In the Jan. 27 issue of Science, the researchers describe their analysis of DNA from microbe communities at seven different depths in the tropical Pacific Ocean, from the surface down to 4,000 meters (about 13,000 feet). One of the team's overall goals was to determine how the microbes near the surface are different from those that live thousands of meters down.
The scientists collected water from the open ocean about 100 kilometers (60 miles) north of the island of Oahu. This spot, site of the Hawaii Ocean Time Series station, has been studied continuously for 18 years by one of the co-authors, David Karl of the University of Hawaii. It was chosen because it is far from any terrestrial influences, yet its chemistry and (nonmicrobial) biology are relatively well known.
Still, challenges remained. Because concentrations of microbes were so low in this "oceanic desert" area, the team had to spend five to six hours filtering up to 600 liters (160 gallons) of seawater for each sample to obtain enough microbial DNA for analysis.
What did the researchers find?
Not surprisingly, in samples from the sunlit waters within about 100 meters of the surface, they discovered many microbial DNA sequences that were associated with photosynthesis. This indicates that many microbes in these waters probably use sunlight as a source of energy.
Surface samples also contained microbial DNA associated with movement and propulsion. "This suggests that movement may be especially important for surface-water microbes, perhaps helping them follow chemical gradients or move from food particle to food particle," said DeLong, who has appointments in the Department of Civil and Environmental Engineering (CEE) and in the Biological Engineering Division.
In contrast, DNA from microbes in deeper waters suggests many may survive by attaching to and breaking down particles of organic material. Such particles continually sink down from the surface waters into the deep sea, providing food for many organisms in the form of "marine snow."
Perhaps the most surprising finding was the large amount of DNA that came from viruses, especially in near-surface waters. Since the researchers excluded free-living viruses from their initial sample, they believe that this viral DNA must have come from viruses that had infected living bacteria.
"We're excited about these new views of microbes we and others are developing by analyzing microbial genomes recovered directly from the environment. The approach is really providing new insight into what makes microbes tick in the real world -- how they affect each other and influence their surrounding environment," DeLong said.
DeLong's colleagues from MIT are Penny Chisholm, the Lee and Geraldine Martin Professor of Environmental Studies in CEE and the Department of Biology; CEE postdoctoral associates Tracy Mincer, Steven Hallam (now at the University of British Columbia), Matthew Sullivan and Niels-Ulrik Frigaard (now at the University of Copenhagen); Virginia Rich, a graduate student in the MIT-Woods Hole Oceanographic Institution Joint Program; and CEE research scientist Asuncion Martinez.
Additional authors of the Science paper are from the Monterey Bay Aquarium Research Institute, the University of Hawaii and San Diego State University.
This work was funded by the Gordon and Betty Moore Foundation, the National Science Foundation, and the U.S. Department of Energy Microbial Genomics Program.
A version of this article appeared in MIT Tech Talk on March 8, 2006 (download PDF).