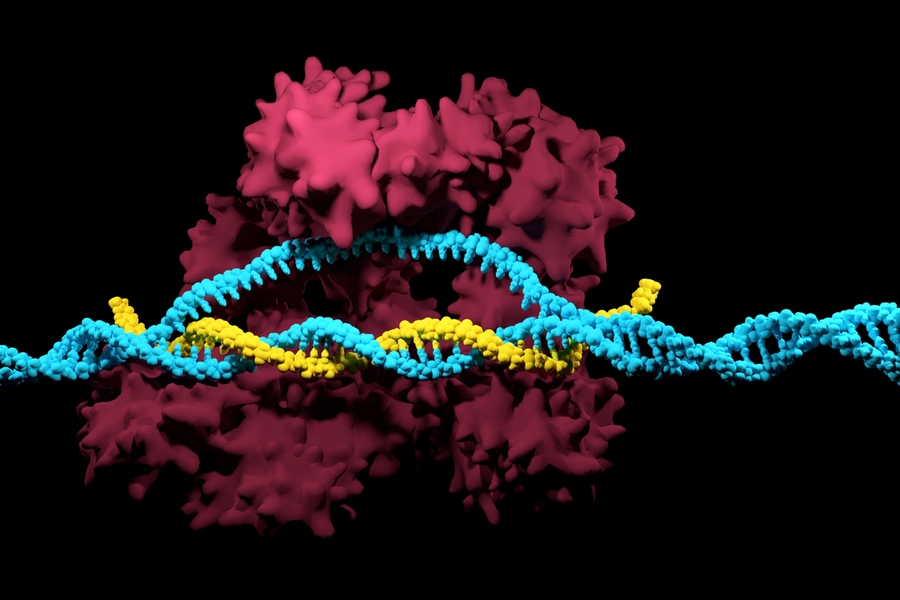
In nature, bacteria use CRISPR as an adaptive immune system to protect themselves against viruses. Over the past decade, scientists have been able to successfully build upon that natural phenomenon with the discovery of CRISPR proteins found in bacteria — the most widely used of which is the Cas9 enzyme. In combination with a guide RNA, Cas9 is able to target, cut, and degrade specific DNA sequences.
With applications ranging from the treatment of genetic diseases to the nutritional potency of agricultural crops, CRISPR has emerged as one of the most promising tools for genome editing. Cas9 enzymes, however, rely on specific DNA ZIP codes to pinpoint where to cut and edit. The most widely-used Cas9 from Streptococcus pyogenes bacteria, SpCas9, requires two “G” nucleotides beside target sites. Less than 10 percent of DNA sequences meet this requirement.
In research published this month in both Nature Biotechnology and Nature Communications, a team of computational biologists in the Media Lab’s Molecular Machines group and the MIT Center for Bits and Atoms have successfully engineered new proteins with enhanced genome editing capabilities, significantly broadening the spectrum of DNA sequences that can be accurately and effectively accessed.
This work was led by Pranam Chatterjee, who recently completed his PhD in media arts and sciences; Noah Jakimo PhD '19, a Media Lab affiliate; and Media Lab Associate Professor Joseph Jacobson, in collaboration with lab members and researchers at the University of Massachusetts Medical School.
These new findings stem from the group's earlier breakthrough work in the computational discovery of Cas9 proteins. The team identified and experimentally characterized the Cas9 from Streptococcus canis bacteria (ScCas9), which, while similar to SpCas9, had the ability to target a much broader range of target DNA sequences. That discovery expanded the number of locations that Cas9 enzymes could target from the original 10 percent of sites on the genome to nearly 50 percent. The team first reported those findings in 2018 in Science Advances.
To improve ScCas9 as a genome editing tool, the scientists computationally identified unique parts from similar Cas9 proteins to engineer an optimized version of ScCas9, which the team has named Sc++.
“Sc++ is the first known enzyme to simultaneously exhibit the three properties deemed essential for effective genome editing: broad targeting capability; robust cutting activity; and minimal errors due to off-targeting,” notes Chatterjee.
Concurrently, the team successfully used their previous SPAMALOT algorithm to discover Streptococcus macacae Cas9 (SmacCas9) that required two “A” nucleotides, rather than two “G”s. Through domain swapping and further engineering, the team presents the new iSpyMac enzyme as one of the first known Cas9 editors not requiring a “G,” enabling targeting of an additional 20 percent of the genome that was previously inaccessible.
"To engineer iSpyMac, we simultaneously made hundreds of changes to SpCas9, knowing even a single change can break it," says Jakimo, the senior author on this second study. "Our success is a testament to the wealth of microbial genomic data that can provide helpful clues about protein function with tools like SPAMALOT."
Erik Sontheimer, professor and vice chair of the RNA Therapeutics Institute at the University of Massachusetts Medical School, and a collaborator on the research, notes the significance of this work. “The fewer targeting limitations we encounter, and the fewer compromises and trade-offs that have to be made between activity and accuracy, the greater the impact that CRISPR genome editing can have on biotechnology and human health. This is why Sc++ and iSpyMac provide such valuable new additions to the CRISPR editing arsenal.”
As labs around the world have already begun to use the enzymes to successfully edit the genomes of various organisms, from rice to rabbits, the next goal for this research will be to develop tools to reach the remaining 30 percent of genome sequences. Chatterjee, in collaboration with the University of Zurich, is looking to unlock the final advances that will allow scientists to access any genomic sequence, and to address any type of gene mutation in the treatment of genetic diseases.
For now, however, as in many labs across the MIT campus, work has pivoted to address the Covid-19 pandemic. By applying computational design principles to engineer proteins that can target and bind to the invading SARS-CoV-2 virus, Chatterjee and the research team at the Media Lab are seeking to create enzymes to rapidly halt the virus, and enable cell recovery.
“We engineer proteins differently," Chatterjee adds. "Our ability to integrate computation and experimentation enables us to refine our algorithms and build impactful tools for a host of applications, from addressing genetic diseases to Covid-19, and beyond.”