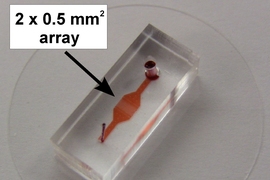
The immune system is a complex network of many different cells working together to defend against invaders. Successfully fighting off an infection depends on the interactions between these cells.
A new device developed by MIT engineers offers a much more detailed picture of that cellular communication. Using this device, which captures pairs of cells and collects data on each as they interact with each other, the researchers have already learned more about how T cells — major players in the immune response — become activated during infection.
The device is based on microfluidic technology developed by Joel Voldman, an MIT professor of electrical engineering and computer science (EECS), in 2009. His team used that earlier version to fuse adult cells with embryonic stem cells, allowing the researchers to observe the genetic reprogramming that occurred in these hybrids.
After that study, immunologists contacted Voldman wondering if the device could be adapted to study immune cells. “A lot of what occurs in the immune system is cells talking to other cells by coming in contact with them,” says Voldman, one of the senior authors of a paper describing the new device in the Jan. 13 issue of Nature Communications.
Voldman and Burak Dura, the paper’s lead author and a graduate student in EECS, spent several years re-engineering the device to get it to work with immune cells, which are much smaller than the cells analyzed in 2009. Hidde Ploegh, an MIT professor of biology and member of the Whitehead Institute for Biomedical Research, is also a senior author of the paper.
Controlled contact
Until now, the most common way to measure interactions between two types of cells was to mix the cells together in a test tube and observe them. However, this approach has limited usefulness because there is no guarantee that each cell is interacting with only one other cell.
“All that uncontrollability makes it hard to interpret the results you get,” Voldman says.
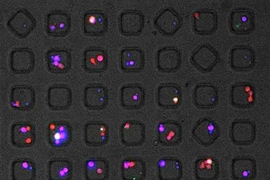
In contrast, Voldman’s device allows for complete control over cell pairings. The device consists of a chip with cell-trapping cups that are strategically arranged to capture and pair up cells. First, type A cells are flowed across the chip in one direction and captured in single-cell traps. Then, the flow of liquid is reversed, drawing the A cells into larger traps located opposite the single cell-traps. When each A cell is in a large trap, B cells are flowed in, and each one joins an A cell in the large traps.
This technique allows the researchers to follow hundreds of cell pairs over time and monitor what is happening in each cell, which has not been possible previously. It also allows them to precisely control the timing of cell interactions.
“We know the exact contact time, and we can keep them in contact as long as they are within the cups,” Dura says. “This allows us to not only measure the single cell parameters but also do measurements of the two cells together and correlate the responses with one another.”
In the new version of the device, the researchers added high-resolution imaging, allowing them to see when cells’ calcium levels fluctuate and when they turn on a type of protein signaling known as phosphorylation.
“This is a very elegant way of doing these experiments,” says Hang Lu, a professor of chemical and biomolecular engineering at the Georgia Institute of Technology who was not involved in the research. “It’s very well-controlled and you know exactly where to look for the cells, and that makes imaging them extremely efficient and high-throughput.”
Launching an immune response
In the Nature Communications paper, Dura worked with Stephanie Dougan, a former postdoc at the Whitehead Institute, to study the interaction between T cells and B cells, which is key to launching an immune response. When B cells encounter viruses or bacteria, they absorb them and display pieces of viral or bacterial proteins (known as antigens) on their cell surfaces. When these B cells encounter T cells with receptors that recognize the antigen, the T cells become activated, provoking them to release cytokines — inflammatory chemicals that control the immune response — or to seek out and destroy infected cells.
Although all of the T cells in this study had identical T cell receptors, the MIT team found that they did not all respond the same way after encountering B cells carrying identical antigens on their surfaces.
Using calcium imaging to measure T cell activation, the researchers found that the initial activation level depends on how much of the antigen is presented. At high levels, most of the cells respond the same way. However, at lower antigen levels, the T cell responses vary greatly. These differences also correlated to differences in T cell cytokine production.
In future studies, the researchers hope to further trace how T cells go through the decision-making process that determines their eventual fates. They also plan to investigate other types of interactions — for example, how immune cells called natural killer cells recognize and destroy cancer cells.
The research was funded by the Singapore-MIT Alliance, the AACR-Pancreatic Cancer Action Network, Janssen Pharmaceuticals, and the Frank Quick Faculty Research Innovation Fellowship.