Engineers at MIT have now built an automated system that can rapidly produce 3-D, micron-resolution images of thousands of zebrafish larvae and precisely analyze their physical traits. The system, described in the Feb. 12 edition of Nature Communications, offers a comprehensive view of how potential drugs affect vertebrates, says Mehmet Fatih Yanik, senior author of the paper.
“Complex processes involving organs cannot be accurately recapitulated in cell culture today. Existing 3-D tissue models are still far too simple to model live animals,” says Yanik, an MIT associate professor of electrical engineering and computer science and biological engineering. “In whole animals, the biology is far more complicated.”
Lead authors of the paper are MIT graduate student Carlos Pardo-Martin and Amin Allalou, a visiting student at MIT. Other authors are MIT senior research scientist Peter Eimon, MIT intern Jaime Medina, and Carolina Wahlby of the Broad Institute.
Zebrafish are genetically similar to humans and have many of the same developmental pathways, so scientists often use them to model human diseases including cancer, diabetes, Parkinson’s disease and autism.
Using the new technology, researchers can grow larvae in tiny wells and flow them through a channel to an imaging platform. Once there, the embryos are rotated and 320 images are taken from different angles, allowing 3-D reconstructions to be made using optical projection tomography (OPT). Getting larvae to the platform takes about 15 seconds, and the imaging takes only 2.5 seconds. This allows hundreds or thousands of larvae to be imaged within hours.
In a 2010 paper, Yanik’s team described the system that transports the embryos to the imaging platform, which they combined with high-resolution two-dimensional imaging. In the latest version, they developed a high-speed OPT imaging technique, which takes hundreds of two-dimensional images and subsequently generates a 3-D image, similar to a CT scan.
They also created a computer algorithm that can measure hundreds of traits and use that information to create a comprehensive phenotype map — the overall description of an organism’s characteristics — for each larva. This enables rapid and detailed studies of how different drugs affect those phenotypes.
“You could probably look at almost any organ or tissue that you’re interested in,” Eimon says. “It gives researchers a way to rapidly measure and quantify and put numbers on the kinds of phenotypes and gene-expression patterns that they’ve been looking at for years and years.”
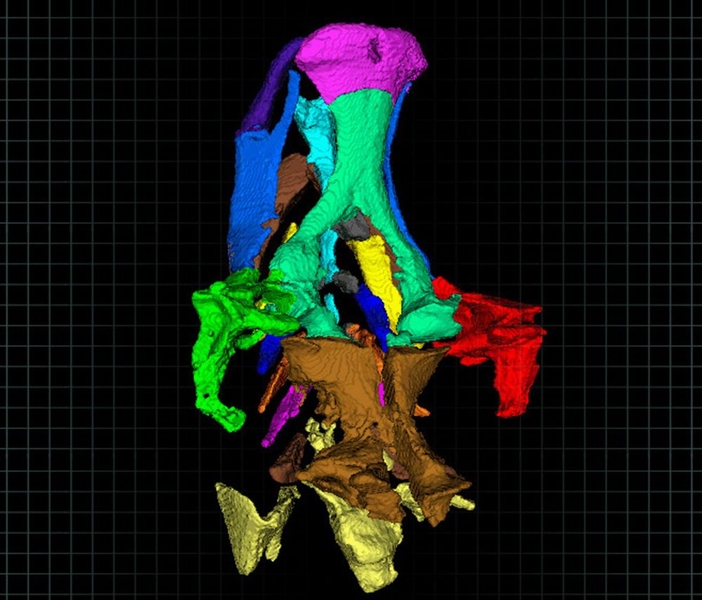
In this study, the researchers focused on the craniofacial skeleton, which is analogous to the human skull. They measured the length and volume of each of the bones that make up this structure, as well as the angles between the bones.
Each embryo was imaged five days after being treated with one of nine different teratogens — drugs that cause developmental abnormalities. The researchers compared their results with the drugs’ known effects and found that they were very consistent. They also obtained high-resolution, 3-D images of the craniofacial skeletons, which are less than a millimeter long.
“Now that we’re able to load the animals, and we can image them really quickly, and we have a way to start looking at the information, the sky’s the limit,” Pardo-Martin says. “What we have to do now is ask the big questions, because the technology has advanced.”
This kind of analysis could be very valuable for drug developers who need to efficiently screen thousands of drug candidates. It could also be used to study hard-to-detect changes in phenotype caused by genetic mutations, says Joseph Fetcho, a professor of neurobiology and behavior at Cornell University.
“A really high-throughput way to assess phenotype is very important for measuring small effects on the development of an organism,” says Fetcho, who was not part of the research team. “You can see what the phenotype looks like in a large population and quantify it in a very rigorous way.”
The software that the researchers wrote to generate the 3-D images is available on their website. The research was funded by the National Institutes of Health, the Packard Award in Science and Engineering, a Sparc Grant from the Broad Institute, and a Howard Hughes Medical Institute Student Fellowship.