Now, researchers in the Computer Science and Artificial Intelligence Laboratory at MIT have developed an algorithm that can analyze information from medical images to identify diseased areas of the brain and their connections with other regions.
The MIT researchers will present the work next month at the International Conference on Medical Image Computing and Computer Assisted Intervention in Nice, France.
The algorithm, developed by Polina Golland, an associate professor of computer science, and graduate student Archana Venkataraman, extracts information from two different types of magnetic resonance imaging (MRI) scans. The first, called diffusion MRI, looks at how water diffuses along the white-matter fibers in the brain, providing insight into how closely different areas are connected to one another. The second, known as functional MRI, probes how different parts of the brain activate when they perform particular tasks, and so can reveal when two areas are active at the same time and are therefore connected.
These two scans alone can produce huge amounts of data on the network of connections in the brain, Golland says. “It’s quite hard for a person looking at all of that data to integrate it into a model of what is going on, because we’re not good at processing lots of numbers.”
So the algorithm first compares all the data from the brain scans of healthy people with those of patients with a particular disease, to identify differences in the connections between the two groups that indicate disruptions caused by the disorder.
However, this step alone is not enough, since much of our understanding of what goes on in the brain concerns the individual regions themselves, rather than the connections between them, making it difficult to integrate this information with existing medical knowledge.
So the algorithm then analyzes this network of connections to create a map of the areas of the brain most affected by the disease. “It is based on the assumption that with any disease you get a small subset of regions that are affected, which then affect their neighbors through this connectivity change,” Golland says. “So our methods extract from the data this set of regions that can explain the disruption of connectivity that we see.”
It does this by hypothesizing, based on an overall map of the connections between each of the regions in the brain, what disruptions in signaling it would expect to see if a particular region were affected. In this way, when the algorithm detects any disruption in connectivity in a particular scan, it knows which regions must have been affected by the disease to create such an impact. “It basically finds the subset of regions that best explains the observed changes in connectivity between the normal control scan and the patient scan,” Golland says.
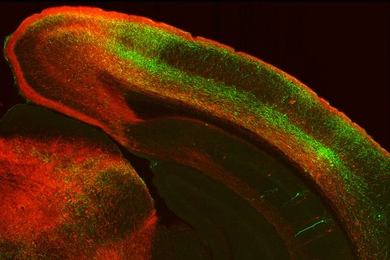
When the team used the algorithm to compare the brain scans of patients with schizophrenia to those of healthy people, they were able to identify three regions of the brain — the right posterior cingulate and the right and left superior temporal gyri — that are most affected by the disease.
In the long term, this could help drug companies develop more effective treatments for the disease that specifically target these regions of the brain, Golland says. In the meantime, by revealing all the different parts of the brain that are affected by a particular disorder, it can help doctors to make sense of how the disease evolves, and why it produces certain symptoms.
Ultimately, the method could also be used to help doctors diagnose patients whose symptoms could represent a number of different disorders, Golland says. By analyzing the patient’s brain scan to pinpoint which regions are affected, it could identify which disorder would create this particular disruption, she says.
In addition to schizophrenia, the researchers, who developed the algorithm alongside Marek Kubicki, associate director of the Psychiatry Neuroimaging Laboratory at Harvard Medical School, are also investigating the possibility of using the method to study Huntington’s disease.
Gregory Brown, associate director of clinical neuroscience at the University of California at San Diego’s Center for Functional MRI, who was not involved in developing the model, plans to use it to study the effects of HIV and drug addiction. “We will use the method to gain a clearer perspective on how HIV infection and methamphetamine dependence disrupts large-scale brain circuitry,” he says.
The method is a critical step away from studying the brain as a collection of localized regions toward a more realistic systems perspective, he says. This should assist the study of disorders such as schizophrenia, neurocognitive impairment and dementia associated with AIDS, and multiple sclerosis, which are best characterized as diseases of brain systems, he says.