MIT chemical engineers have developed a novel way to generate nanoparticles that can recognize specific molecules, opening up a new approach to building durable sensors for many different compounds, among other applications.
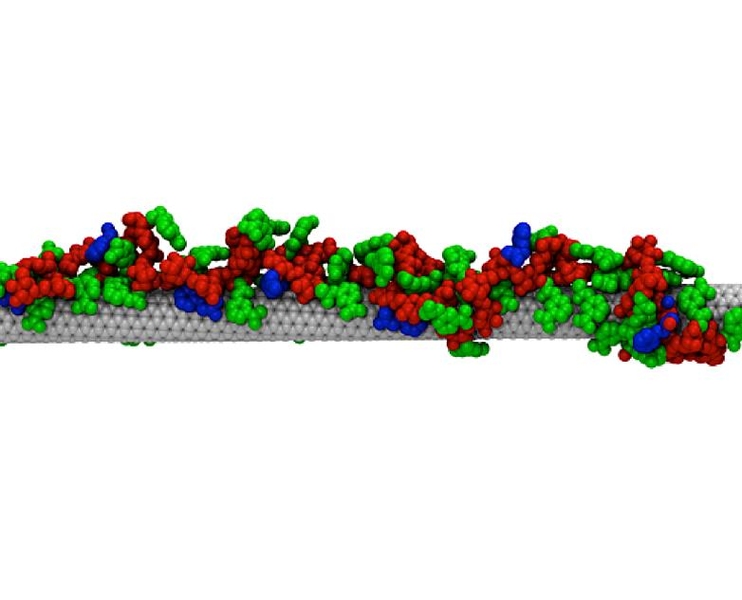
To create these “synthetic antibodies,” the researchers used carbon nanotubes — hollow, nanometer-thick cylinders made of carbon that naturally fluoresce when exposed to laser light. In the past, researchers have exploited this phenomenon to create sensors by coating the nanotubes with molecules, such as natural antibodies, that bind to a particular target. When the target is encountered, the carbon nanotube’s fluorescence brightens or dims.
The MIT team found that they could create novel sensors by coating the nanotubes with specifically designed amphiphilic polymers — polymers that are drawn to both oil and water, like soap. This approach offers a huge array of recognition sites specific to different targets, and could be used to create sensors to monitor diseases such as cancer, inflammation, or diabetes in living systems.
“This new technique gives us an unprecedented ability to recognize any target molecule by screening nanotube-polymer complexes to create synthetic analogs to antibody function,” says Michael Strano, the Carbon P. Dubbs Professor of Chemical Engineering at MIT and senior author of the study, which appears in the Nov. 24 online edition of Nature Nanotechnology.
Lead authors of the paper are recent PhD recipient Jingqing Zhang, postdoc Markita Landry, and former postdocs Paul Barone and Jong-Ho Kim.
Synthetic antibodies
The new polymer-based sensors offer a synthetic design approach to the production of molecular recognition sites — enabling, among other applications, the detection of a potentially infinite library of targets. Moreover, this approach can provide a more durable alternative to coating sensors such as carbon nanotubes with actual antibodies, which can break down inside living cells and tissues. Another family of commonly used recognition molecules are DNA aptamers, which are short pieces of DNA that interact with specific targets, depending on the aptamer sequence. However, there are not aptamers specific to many of molecules that one might want to detect, Strano says.
In the new paper, the researchers describe molecular recognition sites that enable the creation of sensors specific to riboflavin, estradiol (a form of estrogen), and L-thyroxine (a thyroid hormone), but they are now working on sites for many other types of molecules, including neurotransmitters, carbohydrates, and proteins.
Their approach takes advantage of a phenomenon that occurs when certain types of polymers bind to a carbon nanotube. These polymers, known as amphiphilic, have both hydrophobic and hydrophilic regions. These polymers are designed and synthesized such that when the polymers are exposed to carbon nanotubes, the hydrophobic regions latch onto the tubes like anchors and the hydrophilic regions form a series of loops extending away from the tubes.
These loops form a new layer surrounding the nanotube, known as a corona. The MIT researchers found that the loops within the corona are arranged very precisely along the tube, and the spacing between the anchors determines which target molecule will be able to wedge into the loops and alter the carbon nanotube’s fluorescence.
Molecular interactions
What is unique about this approach, the researchers say, is that the molecular recognition could not be predicted by looking at the structure of the target molecule and the polymer before it attaches to the nanotube.
“The idea is that a chemist could not look at the polymer and understand why this would recognize the target, because the polymer itself can’t selectively recognize these molecules. It has to adsorb onto the nanotube and then, by having certain sections of the polymer exposed, it forms a binding site,” Strano says.
Laurent Cognet, a senior scientist at the Institute of Optics at the University of Bordeaux, says this approach should prove useful for many applications requiring reliable detection of specific molecules.
“This new concept, being based on the molecular recognition from the adsorbed phase itself, does not require the use of antibodies or equivalent molecules to achieve specific molecule recognitions and thus provides a promising alternative route for ‘on demand’ molecular sensing,” says Cognet, who was not part of the research team.
The researchers used an automated, robot-assisted trial and error procedure to test about 30 polymer-coated nanotubes against three dozen possible targets, yielding three hits. They are now working on a way to predict such polymer-nanotube interactions based on the structure of the corona layers, using data generated from a new type of microscope that Landry built to image the interactions between the carbon nanotube coronas and their targets.
“What’s happening to the polymer and the corona phase has been a bit of a mystery, so this is a step forward in getting more data to address the problem of how to design a target for a specific molecule,” Landry says.
The research was funded by the National Science Foundation and the Army Research Office through MIT’s Institute for Soldier Nanotechnologies.
To create these “synthetic antibodies,” the researchers used carbon nanotubes — hollow, nanometer-thick cylinders made of carbon that naturally fluoresce when exposed to laser light. In the past, researchers have exploited this phenomenon to create sensors by coating the nanotubes with molecules, such as natural antibodies, that bind to a particular target. When the target is encountered, the carbon nanotube’s fluorescence brightens or dims.
The MIT team found that they could create novel sensors by coating the nanotubes with specifically designed amphiphilic polymers — polymers that are drawn to both oil and water, like soap. This approach offers a huge array of recognition sites specific to different targets, and could be used to create sensors to monitor diseases such as cancer, inflammation, or diabetes in living systems.
“This new technique gives us an unprecedented ability to recognize any target molecule by screening nanotube-polymer complexes to create synthetic analogs to antibody function,” says Michael Strano, the Carbon P. Dubbs Professor of Chemical Engineering at MIT and senior author of the study, which appears in the Nov. 24 online edition of Nature Nanotechnology.
Lead authors of the paper are recent PhD recipient Jingqing Zhang, postdoc Markita Landry, and former postdocs Paul Barone and Jong-Ho Kim.
Synthetic antibodies
The new polymer-based sensors offer a synthetic design approach to the production of molecular recognition sites — enabling, among other applications, the detection of a potentially infinite library of targets. Moreover, this approach can provide a more durable alternative to coating sensors such as carbon nanotubes with actual antibodies, which can break down inside living cells and tissues. Another family of commonly used recognition molecules are DNA aptamers, which are short pieces of DNA that interact with specific targets, depending on the aptamer sequence. However, there are not aptamers specific to many of molecules that one might want to detect, Strano says.
In the new paper, the researchers describe molecular recognition sites that enable the creation of sensors specific to riboflavin, estradiol (a form of estrogen), and L-thyroxine (a thyroid hormone), but they are now working on sites for many other types of molecules, including neurotransmitters, carbohydrates, and proteins.
Their approach takes advantage of a phenomenon that occurs when certain types of polymers bind to a carbon nanotube. These polymers, known as amphiphilic, have both hydrophobic and hydrophilic regions. These polymers are designed and synthesized such that when the polymers are exposed to carbon nanotubes, the hydrophobic regions latch onto the tubes like anchors and the hydrophilic regions form a series of loops extending away from the tubes.
These loops form a new layer surrounding the nanotube, known as a corona. The MIT researchers found that the loops within the corona are arranged very precisely along the tube, and the spacing between the anchors determines which target molecule will be able to wedge into the loops and alter the carbon nanotube’s fluorescence.
Molecular interactions
What is unique about this approach, the researchers say, is that the molecular recognition could not be predicted by looking at the structure of the target molecule and the polymer before it attaches to the nanotube.
“The idea is that a chemist could not look at the polymer and understand why this would recognize the target, because the polymer itself can’t selectively recognize these molecules. It has to adsorb onto the nanotube and then, by having certain sections of the polymer exposed, it forms a binding site,” Strano says.
Laurent Cognet, a senior scientist at the Institute of Optics at the University of Bordeaux, says this approach should prove useful for many applications requiring reliable detection of specific molecules.
“This new concept, being based on the molecular recognition from the adsorbed phase itself, does not require the use of antibodies or equivalent molecules to achieve specific molecule recognitions and thus provides a promising alternative route for ‘on demand’ molecular sensing,” says Cognet, who was not part of the research team.
The researchers used an automated, robot-assisted trial and error procedure to test about 30 polymer-coated nanotubes against three dozen possible targets, yielding three hits. They are now working on a way to predict such polymer-nanotube interactions based on the structure of the corona layers, using data generated from a new type of microscope that Landry built to image the interactions between the carbon nanotube coronas and their targets.
“What’s happening to the polymer and the corona phase has been a bit of a mystery, so this is a step forward in getting more data to address the problem of how to design a target for a specific molecule,” Landry says.
The research was funded by the National Science Foundation and the Army Research Office through MIT’s Institute for Soldier Nanotechnologies.