Scientists have long known that DNA is arranged in a double helix. But if the double helix did not fold further, each cell's genome would be two meters long — far too large to fit into the nucleus of a human cell, which is about a hundredth of a millimeter in diameter.
A new paper from scientists at MIT, the Broad Institute of Harvard and MIT, University of Massachusetts Medical School and Harvard University reveals the three-dimensional structure of the human genome and answers the thorny question of how each of our cells stows some three billion base pairs of DNA.
The work, reported in this week's issue of Science, may also explain how cells control which stretches of DNA are transcribed and which remain silent. Furthermore, the new technique could allow researchers to study how gene expression changes as cells develop or become cancerous, says Thomas Tullius, professor of chemistry at Boston University, who was not part of the research team.
"It's a whole new view of the chromosome and its place in the cell, and it's a view we've never had before," says Tullius, who studies the structure of DNA.
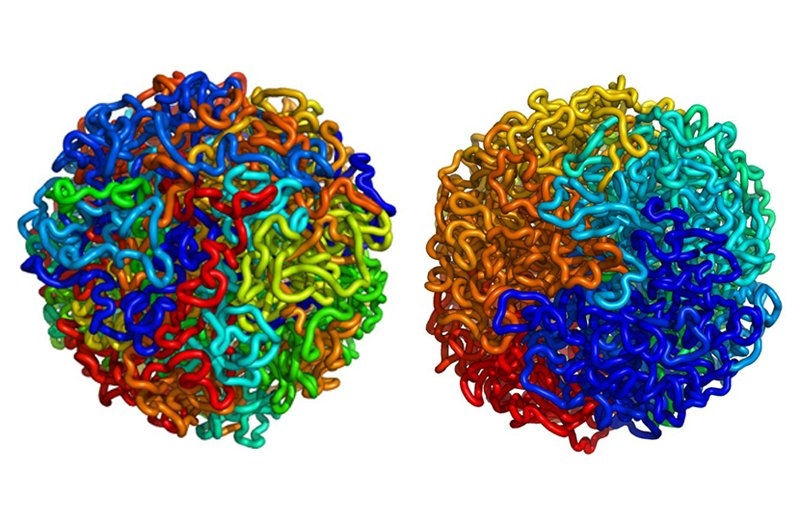
The new structural data reveal that the human genome is organized into two separate compartments, keeping active genes accessible while sequestering unused DNA in a denser storage compartment. Each chromosome alternates between regions of active, gene-rich DNA and inactive, gene-poor stretches.
Collaborating with physicists at MIT, genome scientists established that the genome adopts an unusual organization known in mathematics as a fractal. This architecture, called a "fractal globule," enables the cell to pack DNA incredibly tightly while avoiding the knots and tangles that might interfere with the cell's ability to read its own genome. Moreover, the DNA can easily unfold and refold during gene activation, gene repression and cell replication.
"Nature's devised a stunningly elegant solution to storing information — a super-dense, knot-free structure," says senior author Eric Lander, director of the Broad Institute, who is also professor of biology at MIT and professor of systems biology at Harvard Medical School.
Theoretical biophysicist Alexander Grosberg of New York University originally proposed a fractal globule structure for DNA in 1993. "Now it is beautifully confirmed, which is very exciting," Grosberg says.
Form follows function
Just as the 1953 discovery of the DNA double helix by James Watson and Francis Crick revealed how genetic information is stored and copied, the discovery of DNA's 3-D structure offers new insights into how cells control which sections of DNA are translated into proteins.
"It's very suggestive as to how basic cellular processes are taking place, in terms of information storage and retrieval," says co-first author of the Science paper Erez Lieberman-Aiden, a graduate student in the Harvard-MIT Division of Health Sciences and Technology (HST) and a researcher in Lander's laboratory. "It gives us a lot of ideas about how genes are turned on and off."
For example, computer simulations in the lab of MIT physicist (and HST associate professor) Leonid Mirny demonstrated that sections of a fractal globule structure can be easily opened up by chemical modification, suggesting that cells could use such modifications to control transcription of related genes located near each other.
In the past, many scientists had thought that DNA was compressed into a different architecture called an "equilibrium globule," a configuration that is problematic because it can become densely knotted and does not easily open up.
Key to deciphering the genome's structure was the development of the new Hi-C technique, which permits genome-wide analysis of the proximity of individual genes. The scientists first used formaldehyde to link together DNA strands that are nearby in the cell's nucleus. They then determined the identity of the neighboring segments by shredding the DNA into many tiny pieces, attaching the linked DNA into small loops, and performing massively parallel DNA sequencing.
Lieberman-Aiden observed that the data suggest a fractal globule. He then teamed up with Mirny and Mirny's student Maxim Imakaev to confirm his hypothesis and demonstrate conclusively that the Hi-C data matched fractal globule behavior. Computer simulations further helped to reveal biologically important features of such a DNA architecture.
In future experiments, the researchers hope to follow the development of stem cells into mature cell types such as kidney cells, says Lieberman-Aiden. "We want to understand how that process takes place, because it clearly involves some 3-D remodeling of the nucleus."
(Material from a press release issued by Harvard University and the UMass Medical School was used in this story.)
A new paper from scientists at MIT, the Broad Institute of Harvard and MIT, University of Massachusetts Medical School and Harvard University reveals the three-dimensional structure of the human genome and answers the thorny question of how each of our cells stows some three billion base pairs of DNA.
The work, reported in this week's issue of Science, may also explain how cells control which stretches of DNA are transcribed and which remain silent. Furthermore, the new technique could allow researchers to study how gene expression changes as cells develop or become cancerous, says Thomas Tullius, professor of chemistry at Boston University, who was not part of the research team.
"It's a whole new view of the chromosome and its place in the cell, and it's a view we've never had before," says Tullius, who studies the structure of DNA.
The new structural data reveal that the human genome is organized into two separate compartments, keeping active genes accessible while sequestering unused DNA in a denser storage compartment. Each chromosome alternates between regions of active, gene-rich DNA and inactive, gene-poor stretches.
Collaborating with physicists at MIT, genome scientists established that the genome adopts an unusual organization known in mathematics as a fractal. This architecture, called a "fractal globule," enables the cell to pack DNA incredibly tightly while avoiding the knots and tangles that might interfere with the cell's ability to read its own genome. Moreover, the DNA can easily unfold and refold during gene activation, gene repression and cell replication.
"Nature's devised a stunningly elegant solution to storing information — a super-dense, knot-free structure," says senior author Eric Lander, director of the Broad Institute, who is also professor of biology at MIT and professor of systems biology at Harvard Medical School.
Theoretical biophysicist Alexander Grosberg of New York University originally proposed a fractal globule structure for DNA in 1993. "Now it is beautifully confirmed, which is very exciting," Grosberg says.
Form follows function
Just as the 1953 discovery of the DNA double helix by James Watson and Francis Crick revealed how genetic information is stored and copied, the discovery of DNA's 3-D structure offers new insights into how cells control which sections of DNA are translated into proteins.
"It's very suggestive as to how basic cellular processes are taking place, in terms of information storage and retrieval," says co-first author of the Science paper Erez Lieberman-Aiden, a graduate student in the Harvard-MIT Division of Health Sciences and Technology (HST) and a researcher in Lander's laboratory. "It gives us a lot of ideas about how genes are turned on and off."
For example, computer simulations in the lab of MIT physicist (and HST associate professor) Leonid Mirny demonstrated that sections of a fractal globule structure can be easily opened up by chemical modification, suggesting that cells could use such modifications to control transcription of related genes located near each other.
In the past, many scientists had thought that DNA was compressed into a different architecture called an "equilibrium globule," a configuration that is problematic because it can become densely knotted and does not easily open up.
Key to deciphering the genome's structure was the development of the new Hi-C technique, which permits genome-wide analysis of the proximity of individual genes. The scientists first used formaldehyde to link together DNA strands that are nearby in the cell's nucleus. They then determined the identity of the neighboring segments by shredding the DNA into many tiny pieces, attaching the linked DNA into small loops, and performing massively parallel DNA sequencing.
Lieberman-Aiden observed that the data suggest a fractal globule. He then teamed up with Mirny and Mirny's student Maxim Imakaev to confirm his hypothesis and demonstrate conclusively that the Hi-C data matched fractal globule behavior. Computer simulations further helped to reveal biologically important features of such a DNA architecture.
In future experiments, the researchers hope to follow the development of stem cells into mature cell types such as kidney cells, says Lieberman-Aiden. "We want to understand how that process takes place, because it clearly involves some 3-D remodeling of the nucleus."
(Material from a press release issued by Harvard University and the UMass Medical School was used in this story.)