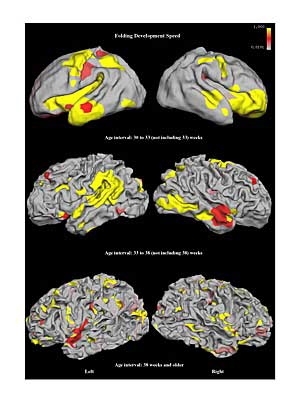
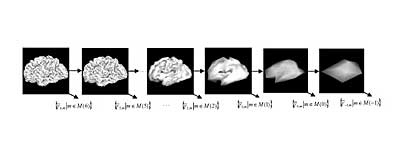
Large mammals--humans, monkeys, and even cats--have brains with a somewhat mysterious feature: The outermost layer has a folded surface. Understanding the functional significance of these folds is one of the big open questions in neuroscience.
Now a team led by MIT, Massachusetts General Hospital and Harvard Medical School researchers has developed a tool that could aid such studies by helping researchers "see" how those folds develop and decay in the cerebral cortex.
By applying computer graphics techniques to brain images collected using magnetic resonance (MR) imaging, they have created a set of tools for tracking and measuring these folds over time. Their resulting model of cortical development may serve as a biomarker, or biological indicator, for early diagnosis of neurological disorders such as autism.
The researchers describe their model and analysis in the April issue of IEEE Transactions on Medical Imaging.
Peng Yu, a graduate student in the Harvard-MIT Division of Health Sciences and Technology (HST), is first author on the paper. The work was led by co-author Bruce Fischl, associate professor of radiology at Harvard Medical School, research affiliate with the MIT Computer Science and Artificial Intelligence Laboratory (CSAIL) and HST, and director of the computational core at the HST Martinos Center for Biomedical Imaging at Massachusetts General Hospital (MGH).
The team started with a collection of MR images from 11 developing brains, provided by Ellen Grant, chief of pediatric radiology at MGH and the Martinos Center. Of the subjects scanned, eight were newborn, mostly premature babies ranging from about 30 to 40 weeks of gestational age, and three were from children aged two, three and seven years. Grant scanned these infants and children to assess possible brain injury and found no neural defects. Later, she also consulted with Fischl's team to ensure that their analyses made sense clinically.
"We can't open the brain and see by eye, but the cool thing we can do now is see through the MR machine," a technology that is much safer than earlier techniques such as X-ray imaging, said Yu.
The first step in analyzing these images is to align their common anatomical structures, such as the "central sulcus," a fold that separates the motor cortex from the somatosensory cortex. Yu applied a technique developed by Fischl to perform this alignment.
The second step involves modeling the folds of the brain mathematically in a way that allows the researchers to analyze their changes over time and space.
The original brain scan is then represented computationally with points. Charting each baby's brain requires about 130,000 points per hemisphere. Yu decomposed these points into a representation using just 42 points that shows only the coarsest folds. By adding more points, she created increasingly finer-grained domains of smaller, higher-resolution folds.
Finally, Yu modeled biological growth using a technique recommended by Grant that allowed her to identify the age at which each type of fold, coarse or fine, developed, and how quickly.
She found that the coarse folds, equivalent to the largest folds in a crumpled piece of paper, develop earlier and more slowly than fine-grained folds.
In addition to providing insights into cortical development, the team is now comparing the images to those being collected from patients with autism. "We now have some idea of what normal development looks like. The next step is to see if we can detect abnormal development in diseases like autism by looking at folding differences," said Fischl. This tool may also be used to shed light on other neurological diseases such as schizophrenia and Alzheimer's disease.
In addition to Yu, Grant and Fischl, co-authors on the paper are postdoctoral associate Yuan Qi and Assistant Professor Polina Golland of CSAIL (Golland also holds an appointment in MIT's Department of Electrical Engineering and Computer Science); Xiao Han of CMS Inc.; Florent Segonne of Certis Laboratory; Rudolph Pienaar, Evelina Busa, Jenni Pacheco and Nikos Makris of the Martinos Center; and Randy L. Buckner of Harvard University and the Martinos Center.
The research was supported by the National Center for Research Resources, the National Institutes of Health, the Washington University Alzheimer's Disease Research Center, and the Mental Illness and Neuroscience Discovery (MIND) Institute. It is part of the National Alliance for Medical Image Computing, funded by the National Institutes of Health.
A version of this article appeared in MIT Tech Talk on April 11, 2007 (download PDF).