CAMBRIDGE, Mass.--The world's smallest photosynthetic organisms, microbes that can turn sunlight and carbon dioxide into living biomass like plants do, will be in the limelight next week. Three international teams of scientists, including a group from MIT, will announce the genetic blueprints for four closely related forms of these organisms, which numerically dominate the phytoplankton of the oceans.
The work will be reported in the August 13 online issues of Nature and the Proceedings of the National Academy of Sciences.
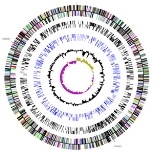
Much like the sequencing of the human genome, the sequencing of the genomes of three strains of Prochlorococcus and one of closely related Synechococcus should crack many mysteries about these organisms--and of phytoplankton in general.
A better understanding of phytoplankton, which play a critical role in the regulation of atmospheric carbon dioxide, will aid studies on global climate change. Further, the "metabolic machinery" of these single-celled organisms could serve as a model for sustainable energy production, as they can turn sunlight into chemical energy.
"The four that have been sequenced represent numerous strains that populate ocean waters and form the base of the food web," said Gabrielle Rocap, lead author of the Nature paper reporting the genetic blueprints for two Prochlorococcus strains. Rocap, a University of Washington assistant professor of oceanography, earned her doctorate from the MIT-Woods Hole Joint Program in 2000.
"A hundred of these organisms can fit end to end across the width of a human hair, but they grow in such abundance that, as small as they are, they at times amount to more than 50 percent of the photosynthetic biomass in the oceans," Rocap said.
ROUGHLY 2,000 GENES
"It behooves us to understand exactly how, with roughly two thousand genes, this tiny cell converts solar energy into living biomass--basic elements into life," said Sallie W. (Penny) Chisholm, the Lee and Geraldine Martin Professor of Environmental Studies in MIT's Department of Civil and Environmental Engineering and Department of Biology.
"These cells are not just some esoteric little creatures," she said. "They dominate the oceans. There are some 100 million Prochlorococcus cells per liter of seawater, for example." Chisholm, the communicating author on the paper by Rocap et al, was part of the team that first described Prochlorococcus in 1988.
Raymond L. Orbach, director of the Department of Energy's Office of Science, which funded part of the research, said, "While many questions remain, it's clear that Prochlorococcus and Synechococcus play an immensely significant role in photosynthetic ocean carbon sequestration. Having the completed genome in hand gives us a first--albeit crude--'parts list' to use in exploring the mechanisms for these and other important processes that could be directly relevant to this critical DOE mission."Synechococcus, a co-inhabitant of ocean waters with Prochlorococcus, has a unique form of motility.
"This research addresses in a concrete way major questions in biological oceanography, at levels finer than the species level," said Phil Taylor, director of the National Science Foundation's biological oceanography program, which also funded part of the research. "The work shows there is a rich and fascinating diversity of physiological capacity and adaptation in the sea, and that this diversity is not always revealed by looking in the microscope."
CLOSE COLLABORATIONS
In the same issue of Nature, a team led by Brian Palenik of the Scripps Institution of Oceanography, part of the University of California at San Diego, will report the sequence of Synechococcus.
The Prochlorococcus and Synechococcus teams collaborated closely. "We learned a tremendous amount working together," said Palenik (who earned his Ph.D. from the MIT-Woods Hole Joint Program in 1989). "By coming at it from different perspectives, we were able to see common themes in how these organisms adapted to the open ocean."
Another paper, written by a team led by Frederick Partensky of the Centre National de la Recherche Scientifique's Station Biologique de Roscoff, describes the genome of a third strain of Prochlorococcus and will be published August 13 in the Proceedings of the National Academy of Sciences.
COMPARATIVE GENOMICS
The work of all three teams "will allow us to better understand what differentiates the ecology of these closely related organisms through comparative genomics," said Chisholm.
The paper by Rocap et al is a kind of case study for how this might work. The scientists report the genetic sequences for two different Prochlorococcus strains, then go on to compare the two. The resulting analysis "reveals many of the genetic foundations for the observed differences in [the two strains'] physiologies and vertical niche partitioning," the authors report. The latter refers to each strain's slightly different ecological niche--they thrive at different depths in the ocean's surface waters.
Chisholm emphasized that "we still don't know the functions of nearly half of these organisms' genes. We're excited about unveiling those functions--particularly for those genes that are unique to the different strains--because they'll alert us to key factors important in regulating marine productivity (photosynthesis) and plankton diversity."
"The idea is to let the organisms tell us what dimensions of their environment are important in determining their distribution and abundance. And this will become easier and easier as the genomes of additional strains are sequenced, and the functions of the genes are understood."
Concluded Rocap, "Right now we don't even know the range of diversity that exists. We've just had a glimpse of the different genome types that are out there."
This research was sponsored by the Department of Energy, with additional support from the Seaver Foundation, the National Science Foundation, the Israel-U.S. Binational Science Foundation and FP5-Margenes.
At MIT, the work is part of the Earth Systems Initiative, organized in November 2002 "to advance our fundamental understanding of how the Earth works," said Chisholm, a co-director. Data from ESI will ultimately help monitor the planet's vital signs, predict the effects of future human activities and otherwise contribute to responsible stewardship of the planet.
In addition to Chisholm, the MIT authors on the Rocap et. al. Nature paper are: Graduate Student Maureen Coleman, Postdoctoral Associate Zachary I. Johnson, Postdoctoral Associate Debbie Lindell, and Postdoctoral Fellow Erik R. Zinser of the Department of Civil and Environmental Engineering; Stephanie L. Shaw (now at the University of California, Berkeley); Graduate Students Matthew B. Sullivan and Andrew Tolonen of the MIT-Woods Hole Joint Program, and Research Scientist Claire S. Ting (now at Williams College).
Other authors of the Nature paper are from the following institutions: Oak Ridge National Laboratory, the Joint Genome Institute, Lawrence Livermore National Laboratory, the University of Washington, Humboldt University in Germany, the Interuniversity Institute of Marine Science in Israel and the Woods Hole Oceanographic Institution.