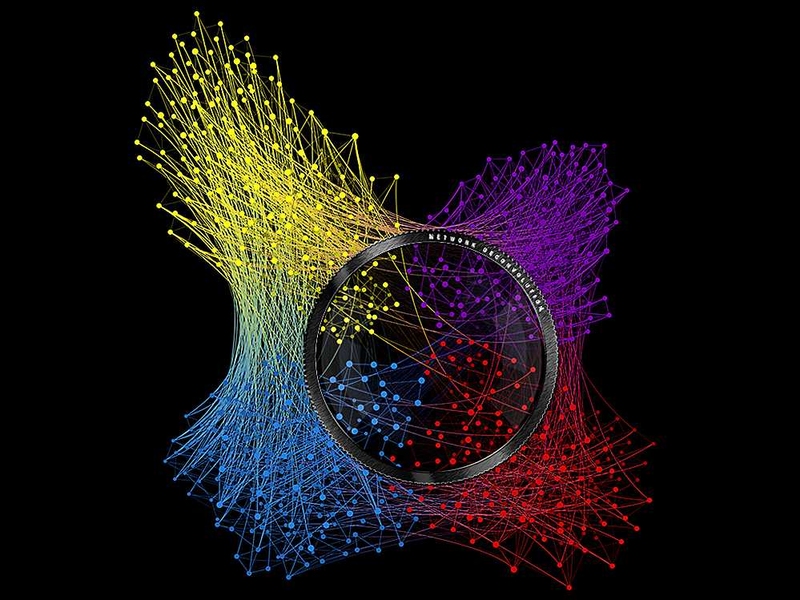
Network science, the study of complex interconnected systems, has grown over the past few years as it has become pivotal in understanding a wide variety of fields ranging from molecular and cell biology to social and information sciences and big data. By studying the structure and connectivity patterns of a network, researchers are able to gain insight into how different variables within a network are related and the strength of individual relationships, as well as make predictions about emerging network properties.
In engineered networks, the connections between elements are directly observable. For most social, biological and information networks, the connections are only inferred based on the behavior of elements in the system across time or conditions.
“The challenge is that some elements may appear correlated in their activity patterns solely because they are both connected to a third party,” says Manolis Kellis, an associate professor of computer science at MIT. “For example, if my son comes to several genomics conferences with me, it may seem like he is part of the genomics community, but the link is only indirect.”
These indirect links have plagued the field of network science since its inception, and as data mining capabilities are able to capture increasingly subtle interactions in increasingly dense networks, the challenge is only greater. Removing indirect effects seems almost impossible, as they are deeply engrained in any network inferred by observing behavioral patterns.
In a new paper appearing in the August edition of Nature Biotechnology, Kellis and colleagues from the MIT Computer Science and Artificial Intelligence Lab (CSAIL), the MIT Research Laboratory of Electronics (RLE), and the Broad Institute, describe a new algorithm that can infer direct dependencies in a network.
“We developed a new approach for distinguishing which interactions are direct and which are redundant by leveraging connectivity patterns from the complete network,” says Soheil Feizi, lead author of the paper and a graduate student at CSAIL and RLE. “We applied it to gene networks, protein folding, and co-authorship social networks, but our method is general and applicable to many other network science problems.”
During their research, the team recognized that a network inferred from real-world observations is in fact a transformation of a true, but hidden, underlying network. The observed links represent the sum, or ‘convolution’, of direct links and their infinitely many echoes over all paths in the network, each with diminishing effects. With this realization, they sought to mathematically reverse the effects of network convolution, by exploiting key properties of matrix decomposition and infinite sums.
They expressed the matrix representing all element correlations as a function of its principal components, and corresponding weights for each component. This transformation allowed the researchers to isolate and remove the effects of network convolution on each component separately, and rebuild the original true network, thus distinguishing the direct connections likely responsible for all observed dependencies.
The technique is similar to trying to distinguish between water and alcohol mixed together in a glass. In liquid form, the two are indistinguishable to the human eye. If frozen, it is immediately apparent which liquid is water and which is alcohol, as the water will freeze and the alcohol will not.
“Transforming the network to a spectral space reveals direct links in a clearer manner,” Feizi says. “Spectral approaches use global information from the complete structure of the network, and are thus much more powerful than previous methods that only looked at local properties to recognize indirect links.”
Most techniques for inferring connections within a network are typically computationally intensive and time-consuming and only applicable to a limited number of situations. The technique developed by the MIT researchers can be applied to a wide variety of areas, from studying gene regulatory networks, to social and online networks and other information sciences, providing insight on disease pathways, social interactions and other diverse fields.
"The technique readily reveals how local interactions affect global behavior,” says Muriel Medard, co-author of the study and a professor in RLE. "It is a versatile analytical tool that can be applied to networks of arbitrary dimension, and fits in a more general framework of applying information theory and linear algebra techniques to real-world problems, with many possible modifications.”
“Introducing such a foundational operation on networks seems surprising in this day and age,” Kellis says. “However, network science is still a young field, and with new insights coming from diverse domains, we expect many such additional surprises to emerge with broad impact in our increasingly interconnected world.”
In engineered networks, the connections between elements are directly observable. For most social, biological and information networks, the connections are only inferred based on the behavior of elements in the system across time or conditions.
“The challenge is that some elements may appear correlated in their activity patterns solely because they are both connected to a third party,” says Manolis Kellis, an associate professor of computer science at MIT. “For example, if my son comes to several genomics conferences with me, it may seem like he is part of the genomics community, but the link is only indirect.”
These indirect links have plagued the field of network science since its inception, and as data mining capabilities are able to capture increasingly subtle interactions in increasingly dense networks, the challenge is only greater. Removing indirect effects seems almost impossible, as they are deeply engrained in any network inferred by observing behavioral patterns.
In a new paper appearing in the August edition of Nature Biotechnology, Kellis and colleagues from the MIT Computer Science and Artificial Intelligence Lab (CSAIL), the MIT Research Laboratory of Electronics (RLE), and the Broad Institute, describe a new algorithm that can infer direct dependencies in a network.
“We developed a new approach for distinguishing which interactions are direct and which are redundant by leveraging connectivity patterns from the complete network,” says Soheil Feizi, lead author of the paper and a graduate student at CSAIL and RLE. “We applied it to gene networks, protein folding, and co-authorship social networks, but our method is general and applicable to many other network science problems.”
During their research, the team recognized that a network inferred from real-world observations is in fact a transformation of a true, but hidden, underlying network. The observed links represent the sum, or ‘convolution’, of direct links and their infinitely many echoes over all paths in the network, each with diminishing effects. With this realization, they sought to mathematically reverse the effects of network convolution, by exploiting key properties of matrix decomposition and infinite sums.
They expressed the matrix representing all element correlations as a function of its principal components, and corresponding weights for each component. This transformation allowed the researchers to isolate and remove the effects of network convolution on each component separately, and rebuild the original true network, thus distinguishing the direct connections likely responsible for all observed dependencies.
The technique is similar to trying to distinguish between water and alcohol mixed together in a glass. In liquid form, the two are indistinguishable to the human eye. If frozen, it is immediately apparent which liquid is water and which is alcohol, as the water will freeze and the alcohol will not.
“Transforming the network to a spectral space reveals direct links in a clearer manner,” Feizi says. “Spectral approaches use global information from the complete structure of the network, and are thus much more powerful than previous methods that only looked at local properties to recognize indirect links.”
Most techniques for inferring connections within a network are typically computationally intensive and time-consuming and only applicable to a limited number of situations. The technique developed by the MIT researchers can be applied to a wide variety of areas, from studying gene regulatory networks, to social and online networks and other information sciences, providing insight on disease pathways, social interactions and other diverse fields.
"The technique readily reveals how local interactions affect global behavior,” says Muriel Medard, co-author of the study and a professor in RLE. "It is a versatile analytical tool that can be applied to networks of arbitrary dimension, and fits in a more general framework of applying information theory and linear algebra techniques to real-world problems, with many possible modifications.”
“Introducing such a foundational operation on networks seems surprising in this day and age,” Kellis says. “However, network science is still a young field, and with new insights coming from diverse domains, we expect many such additional surprises to emerge with broad impact in our increasingly interconnected world.”