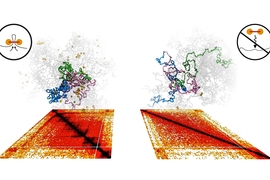
A molecular “motor” that organizes the genome into distinct neighborhoods by forming loops of DNA has been characterized by researchers at MIT and the Pasteur Institute in France.
In a study published in 2016, a team led by Leonid Mirny, a professor of physics in MIT’s Institute for Medical Engineering and Sciences, proposed that molecular motors transform chromosomes from a loosely tangled state into a dynamic series of expanding loops.
The process, known as loop extrusion, is thought to bring regulatory elements together with the genes they control. The team also suggested that DNA is decorated with barriers — akin to stop signs — that limit the process of extrusion.
In this way, loop extrusion divides chromosomes into separate regulatory neighborhoods, known as topologically associating domains (TADs).
However, while the researchers suggested that a ring-like protein complex called cohesin was a likely candidate for these molecular motors, this had yet to be proven.
Now, in a paper published in the journal Nature, a team led by Mirny and Francois Spitz at the Pasteur Institute, have demonstrated that cohesin does indeed play the role of a motor in the loop extrusion process.
“Each of these machines lands on the DNA and starts extruding loops, but there are boundaries on DNA that these motors cannot get through,” Mirny says. “So as a result of this motor activity, the genome is organized into many dynamic loops that do not cross the boundaries, so the genome becomes divided into a series of neighborhoods.”
The researchers also discovered that a different mechanism, that does not use cohesin, is at work organizing active and inactive regions of DNA into separate compartments in the cell’s nucleus.
To determine the role cohesin plays in genome formation, the team first deleted a molecule known as Nipbl, which is responsible for loading cohesin onto DNA.
They then used an experimental technique known as Hi-C, in which parts of DNA that are close to one another in 3-D space are captured and sequenced, in a bid to measure the frequency of physical interactions between different spots along chromosomes.
This technique, which was pioneered by Job Dekker, a professor of biochemistry and molecular pharmacology at the University of Massachusetts Medical Center in Worcester, has previously been used to demonstrate the existence of TADs.
The team first used the Hi-C technique to assess the organization of chromosomes before removing the Nipbl molecule from mice. They then removed the molecule and performed the same measurement again.
They found that the neighborhoods had virtually disappeared.
However, the compartmentalization between active and inactive regions of the genome had become even more marked.
The team believes the cohesin motors allow each gene to reach out to its regulatory elements, which control whether genes should be switched on or off.
What’s more, it appears that the cohesin motors are stopped by another protein, CTCF, which demarcates the boundaries of each neighborhood. In a recent study in the journal Cell, the Mirny lab, in collaboration with researchers at the University of California at San Francisco and the University of Massachusetts Medical School has demonstrated that if this demarcating protein is removed, the borders between neighborhoods disappear, allowing genes in one neighborhood to talk to regulatory elements they should not be talking to in another neighborhood, and leading to misregulation of genes in the cell.
“Cohesin is central for gene regulation, and we emphasize that this is a motor function, so it is not just that they (genes and their regulatory elements) find each other somewhere randomly in space, but they were brought together by this motor activity,” Mirny says.
This paper provides important new molecular insights into the mechanisms by which cells fold their chromosomes, according to Dekker, who was not involved in the current study.
“In this work the Mirny and Spitz labs combine mouse models with genomic approaches to study chromosome folding to reveal that the machine that loads the cohesin complex is critical for TAD formation,” Dekker says. “From this and another previous study, a molecular mechanism is coming into view where TADs form by cohesin and Nipbl-dependent chromatin loop extrusion, which is blocked by sites bound by CTCF.”
The researchers are now attempting to characterize how the absence of the molecular motor would affect gene regulation. They are also carrying out computer simulations in a bid to determine how the cohesin-based loop extrusion takes place at the same time as the genome is undergoing the independent process of segregating into active and inactive compartments.
“It’s like two pianists playing on the same piano,” says Nezar Abdennur, a PhD student in the Mirny lab, who took part in the study alongside fellow PhD student Anton Goloborodko. “They interfere and put constraints on each other, but together they can produce a beautiful piece of music.”
Abdennur and Goloborodko are co-first authors of the paper, along with Wibke Schwarzer of EMBL. The research was supported, in part, by the National Institutes of Health, the National Science Foundation, and the MIT-France MISTI Fund.