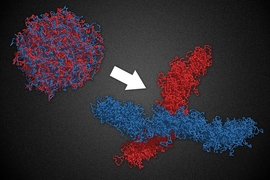
Human cells contain 23 pairs of chromosomes that form a loosely organized cluster in the cell nucleus. When cells divide, they must first condense these chromosomes — each of which when fully extended is a thousand times longer than the cell’s nucleus and physically indistinguishable from the others — into compact structures that can be easily separated and packaged into their offspring.
An MIT-led team has now developed a model that explains how cells handle this difficult task. In computer simulations, the researchers demonstrate that certain molecular “machines” can transform chromosomes from a loosely tangled rope into a series of tiny loops that condense each chromosome and allow it to extricate itself from the others.
Moreover, the researchers demonstrate that a similar model explains how chromosomes are organized when cells are not dividing, and they hypothesize that loop extrusion by molecular motors splits chromosomes into separate domains, helping to control which genes are expressed in a given cell.
This mechanism, outlined in three recent papers published in Cell Reports, eLife, and Biophysical Journal, suggests that chromosome organization relies on proteins that act as molecular motors that pull strands of DNA into progressively larger loops. The MIT team suggests that two proteins thought to function primarily as “staples” that hold DNA together, cohesin and condensin, can also actively manipulate DNA.
“Nobody has ever directly observed this mechanism of loop extrusion. If it exists, it will solve lots of problems,” says Leonid Mirny, a professor of physics in MIT’s Institute for Medical Engineering and Sciences, who led the research. “We will know how chromosomes condense, how they segregate, how genes talk to enhancers. Lots of things can be solved by this mechanism.”
Condensation by loops
This loop extrusion hypothesis was proposed in 2012 by John Marko, a professor of physics and molecular biosciences at Northwestern University, who is an author on the eLife and Biophysical Journal papers. Marko and the MIT researchers, led by graduate student Anton Goloborodko, teamed up to use computer simulations of chromosomes to investigate whether loops could produce the compact chromosomes seen in dividing cells.
From a physics point of view, it was puzzling how loosely organized chromosomes could be condensed into a compact, elongated structure, Mirny says. Most passive condensation strategies, such as simply stapling the chromosomes together, would result in a disordered sphere, he says.
Goloborodko compares the task to trying to respool a tangled fishing line.
“When I was a kid, I used to go fishing with my father, and inevitably, my fishing line would end up tangled into a ball, so that I'd have to spend hours fixing it. Our cells are faced with a similar but much harder task every time they have to divide: They have to nicely spool and separate their chromosomes, but within the tight space of their nuclei. That would be equal to asking someone to untangle and spool a few kilometers of fishing line in the space of a shoe box,” he says.
Using computer simulations that model the dynamics of interactions between chromosomes and proteins, the researchers analyzed what would happen if many proteins acting as molecular motors pulled DNA strands into progressively larger loops. Each of these motors acts independently, with no communication among them. The simulations revealed that this mechanism can successfully condense chromosomes into the distinctive, elongated structures seen when cells divide.
The researchers theorize that in living cells, as the cells enter metaphase — the part of their life cycle where chromosomes condense in preparation for cell division — many molecules of condensin land on the chromosomes and begin the loop extrusion process.
“What we believe is happening is each condensin complex consists of two motors, each moving along the chromosome in the opposite direction,” Mirny says. “Each loop extrusion event is tiny compared to the size of a chromosome. Nevertheless, collectively they can self-organize this chromosome into an early-metaphase, condensed chromosome.”
In human cells, this process usually takes about 20 minutes.
Genetic regulation
In their Cell Reports paper, the MIT researchers put forth a model of how loop extrusion by molecular motors splits chromosomes into smaller domains during interphase — the part of the cell cycle during which cells carry out their particular function, rather than preparing to divide.
This model helps to explain a previous discovery that within regions of chromosomes known as chromosomal domains, genes and regulatory elements interact more with each other than they do with those in neighboring domains. Each chromosome has hundreds of these domains.
Other researchers have shown that these domains are separated by boundaries where many diverse proteins are directly associated with the chromosome. The new MIT research suggests that these boundaries prevent loops from spreading from one region to another, bringing genetic regulatory elements close to genes within the boundaries and preventing them from interacting with genes outside the boundaries.
“It’s a mechanism whereby interactions can be facilitated and also insulated from happening. You can stop certain regulatory elements from talking to certain genes,” says Geoffrey Fudenberg, a fellow at the Center for 3D Structure and Physics of the Genome at MIT and the University of Massachusetts Medical School, who is co-first author of the Cell Reports paper, along with recent MIT PhD recipient Maxim Imakaev, currently a postdoc at Harvard Medical School.
The loops seen during interphase are constantly being formed and then dismantled in a dynamic process. When a cell begins preparing to divide, the researchers believe the boundaries are removed and condensin comes in to form the many more loops needed to condense the chromosomes for cell division.
Although biologists have not demonstrated that condensin and cohesin can act as molecular motors, there is speculation that they can because they are structurally similar to other cellular proteins that function as molecular motors.
“Combined, these papers constitute the most important papers in our field in the last five years,” says Job Dekker, a professor of biochemistry and molecular pharmacology at the University of Massachusetts Medical School, who was not involved in the research. “These papers explain how a very simple method of loop extrusion can explain, to an extraordinary level, the structures of chromosomes in both mitotic and nondividing cells.”
The Mirny and the Marko labs are parts of the Center for 3D Structure and Physics of the Genome, funded by the new National Institutes of Health 4D Nucleome Consortium. The center, co-directed by Dekker and Mirny, brings together genomics, imaging, and computational groups to characterize chromosomal organization in human cells. The team now hopes to generate more data to test the loop extrusion mechanism. Mirny’s lab is also investigating how different types of cells vary in where they place boundaries, which could help explain how different cells express different genes.
The project was started as a part of MIT PRIMES, a high school outreach program. PRIMES participant Carolyn Lu, currently an MIT undergraduate, is an author of the Cell Reports paper. Nezar Abdennur, a graduate student in the Computational and Systems Biology program, is also an author of the Cell Reports paper.