These induced pluripotent stem cells (iPSCs) are usually created by genetically modifying cells to overexpress four genes that make them revert to an immature, embryonic state. However, the procedure works in only a small percentage of cells.
Now, new genetic markers identified by researchers at MIT and the Whitehead Institute could help make that process more efficient, allowing scientists to predict which treated cells will successfully become pluripotent.
The new paper, published in the Sept. 13 online edition of Cell, also identifies new combinations of reprogramming factors that produce iPSCs, according to the researchers.
Led by Rudolf Jaenisch, an MIT professor of biology and member of the Whitehead Institute, the study is the first to examine genetic changes that occur in individual cells as they become pluripotent. Previous studies have only looked at gene-expression changes in large populations of cells — not all of which will actually reprogram — making it harder to pick out genes involved in the process.
“In previous studies, you weren’t able to detect the few cells that expressed predictive pluripotency markers. The really cool part of this study is that you can detect two or three cells that express these important genes early, which has never been done before,” says Dina Faddah, a graduate student in Jaenisch’s lab and one of the paper’s lead authors.
The other lead author is Yosef Buganim, a postdoc at the Whitehead Institute.
Single-cell analysis
In 2007, scientists discovered that adult human cells could be reprogrammed by overexpressing four genes — Oct4, Sox2, c-Myc and Klf4. However, in a population of cells in which those genes are overexpressed, only about 0.1 to 1 percent will become pluripotent.
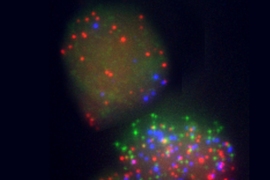
In the new study, Jaenisch’s team reprogrammed mouse embryonic fibroblast cells and then measured their expression of 48 genes known or suspected to be involved in pluripotency at several points during the process. This allowed them to compare gene-expression profiles in cells that became pluripotent, those that did not, and those that were only partially reprogrammed.
Once the reprogramming was complete, which took between 32 and 94 days, the researchers looked for genes expressed only in the cells that ended up becoming pluripotent.
The team identified four genes that were turned on very early — around six days after the reprogramming genes were delivered — in cells that ended up becoming pluripotent: Esrrb, Utf1, Lin28 and Dppa2, which control the transcription of other genes involved in pluripotency.
The researchers also found that several previously proposed markers for pluripotency were active in cells that became only partially programmed, suggesting those markers would not be useful. With their newly discovered markers, “you can eliminate all the colonies that are not completely reprogrammed,” Buganim says. “You don’t want to use partially reprogrammed iPSCs for patient-specific therapies.”
To read cells’ genetic profiles so precisely, the researchers screened for genes using a microfluidic system called Fluidigm, then confirmed their results with a fluorescence imaging technique that can detect single strands of messenger RNA.
Not totally random
The findings also allowed the researchers to develop a new model for how genes interact with each other to steer cells toward pluripotency. Previously, it had been thought that reprogramming was a random process — that is, once the four reprogramming genes were overexpressed, it was a matter of chance whether they would activate the correct genes to make a particular cell pluripotent.
However, the new study reveals that only the earliest phase of the process is random. Once those chance events awaken the cell’s own dormant copy of the Sox2 gene, that gene launches a deterministic pathway that leads to pluripotency.
During the early, random stage, there are probably many ways that Sox2 can be activated, Buganim says. “Different cells will activate Sox2 in different ways,” he says. “As soon as you have a specific combination that allows the activation of Sox2, you are on the way toward full reprogramming.”
The new model also predicted six combinations of factors that could activate Sox2. The researchers tested these combinations in reprogrammed cells and found that they were successful, with varying rates of efficiency.
“It’s a beautiful demonstration of how single-cell gene-expression data can be used for bioinformatics and predicting the gene hierarchy that regulates the reprogramming process,” says Andras Nagy, a senior investigator at the Samuel Lunenfeld Research Institute in Toronto, who was not involved in this research.
Interestingly, the MIT team found combinations that do not include any of the original reprogramming factors. The researchers are now testing their new combinations to see if they produce healthier iPSCs. The most stringent test involves injecting iPSCs into an embryo that cannot give rise to normal cells because it has four sets of chromosomes instead of two. If a healthy animal develops from those cells, it is entirely the product of the iPSCs, demonstrating that the iPSCs were equivalent to embryonic stem cells. Most iPSCs injected into embryos do not pass this test.
The research was funded by the National Institutes of Health and the NIH/NCI Physical Sciences Oncology Center at MIT.