MIT researchers have figured out how bacteria ensure that they respond correctly to hundreds of incoming signals from their environment.
The researchers also successfully rewired the cellular communications pathways that control those responses, raising the possibility of engineering bacteria that can serve as biosensors to detect chemical pollutants. The work is reported in the June 13 issue of Cell.
Led by MIT biology professor Michael Laub, the team studied genomes of nearly 200 bacteria, which can have hundreds of different pathways that respond to different types of external stimuli. Nutrients, antibiotics, temperature or light can evoke a variety of responses, including transcription of particular genes.
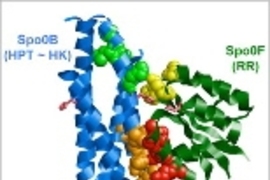
In most cases, the pathways involve two proteins. The first protein, an enzyme known as a histidine kinase, receives the external signal and then activates the second protein, known as a response regulator.
It's critical that each histidine kinase activate only the appropriate response regulator. Different histidine kinases are often very structurally similar, as are the response regulator proteins, so scientists have wondered how cells prevent signals from getting crossed.
"If an organism has tons of this class of signaling pathway, why do we not get a lot of crosstalk?" said Laub. "How does the kinase pick out the right target?"
Based on earlier studies, the MIT researchers theorized that the specificity of the interaction is determined by a subset of amino acids on the histidine kinase and a corresponding subset of amino acids on the response regulator.
To confirm their theory, they looked for patterns of amino acid co-evolution in pairs of histidine kinases and their target response regulators.
Co-evolution occurs when a mutation in one of the two proteins is followed by a secondary mutation in the corresponding amino acid on the other protein, allowing the protein pair to maintain their interaction.
After searching a vast database of nearly 1,300 protein pairs, they identified a small set of co-evolved amino acids. They then confirmed that these amino acids govern signaling specificity by successfully rewiring five of the pathways by mutating the target amino acids.
Such manipulation could allow scientists to engineer bacteria that exhibit novel behavior such as glowing when they detect the presence of a pollutant such as toluene, said Laub.
Lead author of the paper is Jeffrey Skerker, a former MIT postdoctoral associate now at the Broad Institute. Other MIT authors are Barrett Perchuk, technical associate in the Department of Biology, and graduate students Emma Lubin and Orr Ashenberg.
The research was funded by the U.S. Department of Energy and the National Institutes of Health.