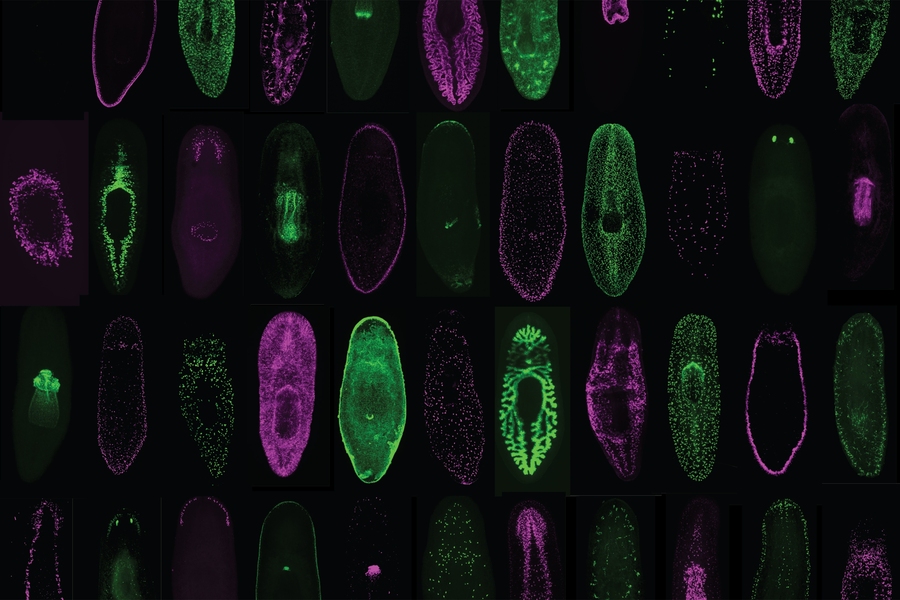
A team at Whitehead Institute and MIT has harnessed single-cell technologies to analyze over 65,000 cells from the regenerative planarian flatworm, Schmidtea mediterranea, revealing the complete suite of actives genes (or “transcriptome”) for practically every type of cell in a complete organism. This transcriptome atlas represents a treasure trove of biological information on planarians, which is the subject of intense study in part because of its unique ability to regrow lost or damaged body parts. As described in the April 19 advance online issue of the journal Science, this new, publicly available resource has already fueled important discoveries, including the identification of novel planarian cell types, the characterization of key transition states as cells mature from one type to another, and the identity of new genes that could impart positional cues from muscles cells — a critical component of tissue regeneration.
“We’re really at the beginning of an amazing era,” says senior author Peter Reddien, a member of Whitehead Institute, professor of biology at MIT, and investigator with the Howard Hughes Medical Institute. “Just as genome sequences became indispensable resources for studying the biology of countless organisms, analyzing the transcriptomes of every cell type will become another fundamental tool — not just for planarians, but for many different organisms.”
The ability to systematically reveal which genes in the genome are active within an individual cell flows from a critical technology known as single-cell RNA sequencing. Recent advances in the technique have dramatically reduced the per-cell cost, making it feasible for a single laboratory to analyze a suitably large number of cells to capture the cell type diversity present in complex, multi-cellular organisms.
Reddien has maintained a careful eye on the technology from its earliest days because he believed it offered an ideal way to unravel planarian biology. “Planarians are relatively simple, so it would be theoretically possible for us to capture every cell type. Yet they still have a sufficiently large number of cells — including types we know little or even nothing about,” he explains. “And because of the unusual aspects of planarian biology — essentially, adults maintain developmental information and progenitor cells that in other organisms might be present transiently only in embryos — we could capture information about mature cells, progenitor cells, and information guiding cell decisions by sampling just one stage, the adult.”
Two and a half years ago, Reddien and his colleagues — led by first author Christopher Fincher, a graduate student in Reddien’s laboratory — set out to apply single-cell RNA sequencing systematically to planarians. The group isolated individual cells from five regions of the animal and gathered data from a total of 66,783 cells. The results include transcriptomes for rare cell types, such as those that comprise on the order of 10 cells out of an adult animal that consists of roughly 500,000 to 1 million cells.
In addition, the researchers uncovered some cell types that have yet to be described in planarians, as well cell types common to many organisms, making the atlas a valuable tool across the scientific community. "We identified many cells that were present widely throughout the animal, but had not been previously identified. This surprising finding highlights the great value of this approach in identifying new cells, a method that could be applied widely to many understudied organisms," Fincher says.
“One main important aspect of our transcriptome atlas is its utility for the scientific community,” Reddien says. “Because many of the cell types present in planarians emerged long ago in evolution, similar cells still exist today in various organisms across the planet. That means these cell types and the genes active within them can be studied using this resource.”
The Whitehead team also conducted some preliminary analyses of their atlas, which they've dubbed “Planarian Digiworm.” For example, they were able to discern in the transcriptome data a variety of transition states that reflect the progression of stem cells into more specialized, differentiated cell types. Some of these cellular transition states have been previously analyzed in painstaking detail, thereby providing an important validation of the team’s approach.
In addition, Reddien and his colleagues knew from their own prior, extensive research that there is positional information encoded in adult planarian muscle — information that is required not only for the general maintenance of adult tissues but also for the regeneration of lost or damaged tissue. Based on the activity pattern of known genes, they could determine roughly which positions the cells had occupied in the intact animal, and then sort through those cells’ transcriptomes to identify new genes that are candidates for transmitting positional information.
“There are an unlimited number of directions that can now be taken with these data,” Reddien says. “We plan to extend our initial work, using further single-cell analyses, and also to mine the transcriptome atlas for addressing important questions in regenerative biology. We hope many other investigators find this to be a very valuable resource, too.”
This work was supported by the National Institutes of Health, the Howard Hughes Medical Institute, and the Eleanor Schwartz Charitable Foundation.