In 1969, scientist Michael Cashel was analyzing the compounds produced by starved bacteria when he noticed two spots appearing on his chromatogram as if by magic. Today, we know one of these “magic spots,” as researchers call them, as guanosine tetraphosphate, or ppGpp for short. We also understand that it is a signaling molecule present in virtually all bacteria, helping tune cell growth and size based on nutrient availability.
And yet, despite decades of study, precisely how ppGpp regulates bacterial growth has remained rather mysterious. Delving further requires a more comprehensive list of the molecules that ppGpp binds to exert its effects.
Now, collaborators from MIT’s departments of Biology and Chemistry have developed a method to do just that, and used their new approach to pinpoint over 50 ppGpp targets in Escherichia coli — roughly half which had not been identified previously. Many of these targets are enzymes required to produce nucleotides, the building blocks of DNA and RNA. During times when the bacteria do not have enough nutrients to grow and divide normally, the researchers propose that ppGpp prevents these enzymes from creating new nucleotides from scratch, helping cells enter a dormant state.
“With small molecules or metabolites like ppGpp, it’s been difficult historically to determine which proteins they bind,” says Michael Laub, a professor of biology, a Howard Hughes Medical Institute investigator, and the senior author of the study. “This has been an intractable problem that’s held the field back for some time, but our new approach allows you to nail down the likely targets in a matter of weeks.”
Postdoc Boyuan Wang is the first author of the study, which appeares in Nature Chemical Biology on Dec. 17.
The magic spot
Since ppGpp was discovered nearly 50 years ago, it has been shown to suppress DNA replication, transcription, translation, and various metabolic pathways. It puts the brakes on cell growth and allows bacteria to persist in the face of starvation, stress, and antibiotics. Its influence over numerous regulatory processes has remained somewhat of a mystery, however — after all, it doesn’t just modulate a single pathway but coordinates multiple operations simultaneously to orchestrate a mass shutdown of the cell.
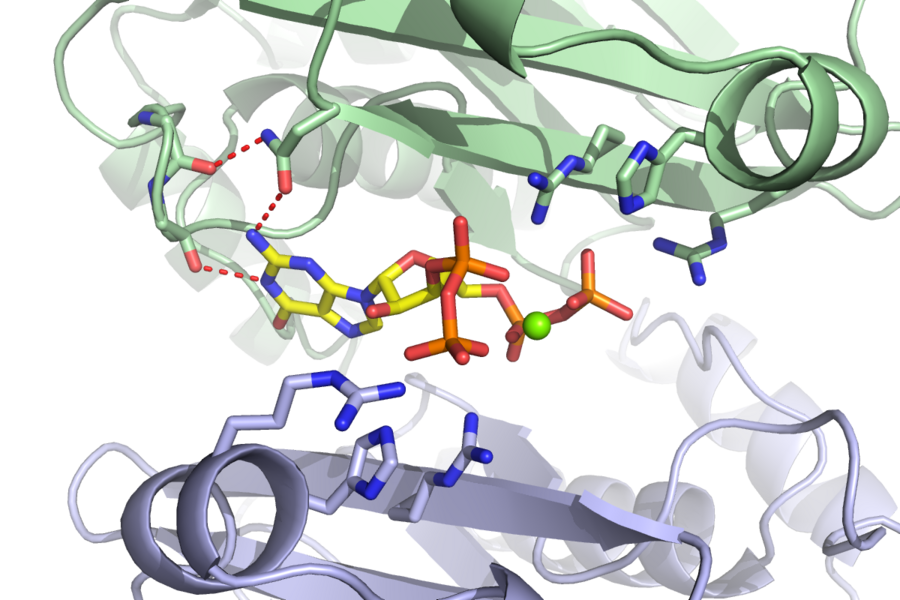
In order to discern which proteins ppGpp binds to effect such widespread change, the researchers built what they call “capture compounds” that contain ppGpp, allowing them to fish out its targets from bacterial extracts. These compounds included a photoreactive crosslinker that latched tightly onto the proteins of interest in the presence of light, and a biotin handle that helped the scientists pull out the proteins to identify them. Most importantly, they were joined to ppGpp in such a way that they wouldn’t interfere with its ability to bind to its targets. This method is more efficient and accurate compared to more traditional means of distinguishing ppGpp targets, which are far more arduous and lack sensitivity.
“Our approach solves these problems because you’re no longer required to do such labor-intensive protocols in order to identify ppGpp targets — and it works even in bacteria beyond E. coli,” says Wang. “Although ppGpp is common among many bacterial species, it seems to exert its effects through different mechanisms, which complicates things. Our capture compounds provide a way to unravel this diversity, and in short order.”
Putting the brakes on nucleotide production
Although the 56 ppGpp targets Wang identified in his screen control a myriad of cellular processes, he homed in on the enzyme PurF — which initiates the biosynthesis of purine nucleotides bearing adenine and guanine bases, also known as A and G.
When bacteria are stressed or starved, they enter a dormant state to survive. But simply curbing translation and transcription is not enough; nucleotides are still being generated and will build up if their synthesis is not put on pause. Cells can build nucleotides in one of two ways: either by salvaging existing materials or starting completely from scratch. PurF kicks off the first step in the latter process leading to the A and G nucleotides. However, when ppGpp binds to PurF, it causes the enzyme to change its shape, which prevents it from doing its job, thus reducing nucleotide production in the cell.
“This is the first time that an enzyme involved in that specific pathway or function has been identified as a ppGpp target,” Wang says. “If you limit the consumption of nucleotides but not their production, the nucleotide pool is going to explode, which isn’t good for the cell. So we’ve shown that ppGpp actually addresses this problem as well.”
In addition to PurF and other enzymes required for nucleotide production, the researchers noticed that ppGpp also binds to many GTPase enzymes involved in translation. This could indicate a failsafe mechanism slowing down translation by striking multiple, similar enzymes in an almost redundant manner in the face of starvation.
As Wang continues to refine his method, he aims to increase its specificity and ensure his capture compounds bind to the exact same proteins they would inside a live cell. He also hopes to screen for ppGpp binding proteins in other bacteria, including pathogens that rely on ppGpp to survive within their hosts and propagate conditions like tuberculosis.
“This is an exciting chemical approach to better understand the function of a long-studied conserved signaling molecule in bacteria,” says Jue Wang, professor of bacteriology at the University of Wisconsin at Madison, who was not involved with the study. “Their findings and techniques are highly relevant to many other bacteria, and will greatly improve knowledge of how bacteria use this critical signaling molecule to mediate everything from surviving in the human gut to causing disease.”
Adds Laub: “We are still discovering new nucleotide-based signaling molecules in bacteria even today, and every single one of them could eventually be derivatized in a similar way to identify their binding partners.”
This research was supported by a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and a grant from the National Institutes of Health.