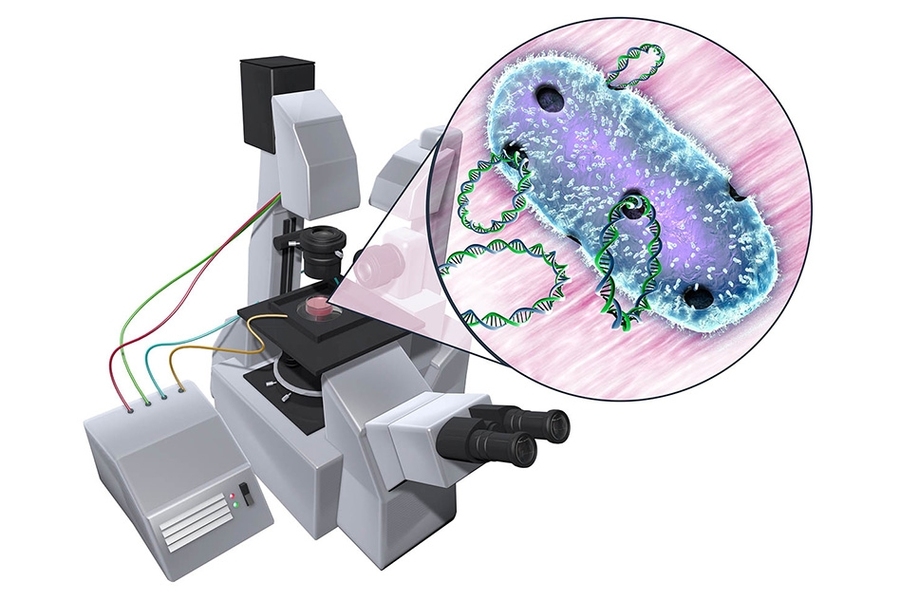
Genetically engineering any organism requires first getting its cells to take in foreign DNA. To do this, scientists often perform a process called electroporation, in which they expose cells to an electric field.
If that field is at just the right magnitude, it will open up pores within the cell membrane, through which DNA can flow. But it can take scientists months or even years to figure out the exact electric field conditions to reversibly unlock a membrane’s pores.
A new microfluidic device developed by MIT engineers may help scientists quickly home in on the electric field “sweet spot” — the range of electric potentials that will harmlessly and temporarily open up membrane pores to let DNA in. In principle, the simple device could be used on any microorganism or cell, significantly speeding up the first step in genetic engineering.
“We’re trying to reduce the amount of experimentation that’s needed,” says Cullen Buie, the Esther and Harold E. Edgerton Associate Professor of mechanical engineering at MIT. “Our big vision for this device and future iterations is to be able to take a process that usually takes months or years, and do it in a day or two.”
Buie and his colleagues, including postdoc Paulo Garcia, graduate student Zhifei Ge, and lecturer Jeffrey Moran, have published their results this week in the journal Scientific Reports.
“A shot in the dark”
Currently, scientists can order various electroporation systems — simple instruments that come with a set of instructions for penetrating an organism’s cell membranes. Each system may include instructions for roughly 100 different organisms, such as strains of bacteria and yeast, each of which requires a unique electric field and set of experimental conditions for permeation. However, Buie says the number of organisms for which these instructions are known is but a fraction of what actually exists in nature.
“There’s a tremendous amount of biodiversity we’re unable to access,” Buie says. “Part of the problem is, we can’t even get the DNA in, much less get it expressed by the organism. And for electroporation, the search for the conditions that might work is like a shot in the dark.”
For electroporation to work, the applied electric field must be strong enough to puncture the membrane temporarily, but not so strong as to do so permanently, which would cause a cell to die.
“It’s like surgery — this is pretty invasive,” Buie says. “There’s a sweet spot between killing them and not affecting them at all, that you need to find to be able to open them reversibly, just enough so that DNA gets in and they reseal on their own.”
Whether an electric field penetrates a membrane also depends on a cell’s surrounding conditions. Scientists have also had to experiment with parameters such as the composition of a cell’s solution and the way in which the electric field is applied.
“For a novel organism, it could take you months or years to develop new conditions so that the cell is happy and will survive the poration process, and it will uptake the DNA,” Buie says.
A range in one
The group’s new microfluidic device may significantly shorten the time it takes to identify these ideal conditions. The device consists of a channel created using soft lithography. The channel narrows in the middle. When an electric field is applied to the device, the channel’s geometry causes the field to exhibit a range of electric potentials, the highest being at the channel’s narrowest region.
The researchers flowed several strains of bacterial cells through the device and exposed the cells to an electric field. They then added a fluorescent marker that lights up in the presence of DNA. If cells were successfully permeated by the electric field, they would let in the fluorescent marker, which would then light up in response to the cell’s own genetic material. To identify the magnitude of the electric potential that was able to open a cell membrane, the researchers simply marked the location of each fluorescent cell along the channel.
“In one experiment, you can test a range of electric fields and get some information almost instantly, in terms of whether there’s been something successful in opening pores,” Buie says. “So now, in your searching process, you don’t need to run a bunch of different experiments and test different electric fields separately. You can do it in one go, and it literally lights up. ”
The researchers successfully permeated strains of E. coli and Mycobacterium smegmatis, a bacterium in the same family as the organism that causes tuberculosis — a family whose membranes, Buie says, are “notoriously difficult” to penetrate.
The group’s first set of experiments involved opening up pores to take in the fluorescent marker — a molecule that is slightly smaller than DNA. The researchers also ran experiments in which they applied an electric field to bacterial cells in the presence of DNA encoded for antibiotic resistance. The team checked that the cells took up the DNA by removing them from the device and growing them on a separate plate with antibiotics — a standard procedure known as a streak test. They found that the cells were able to reproduce — a sign that the DNA was successfully incorporated, and the membranes closed back up.
“At present, only a limited number of cell types can be genetically modified due to limitations in the technologies available for introducing DNA into cells,” says Garcia, who worked on the device as a fellow in MIT’s Translational Fellows Program. “We have developed a microfluidic device that will facilitate genetic engineering of many different cell types. By mediating genetic engineering of novel cell types, this technology will contribute to the areas of drug discovery, regenerative medicine, cancer therapy, and DNA vaccination.”
This research was supported, in part, by DARPA and the National Science Foundation.