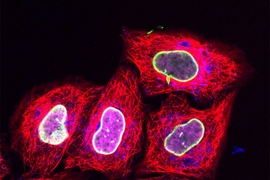
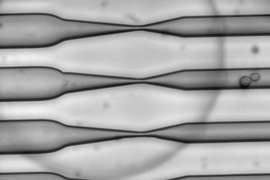
Tagging proteins with a fluorescent label such as green fluorescent protein (GFP) is currently the best way to track specific molecules inside a living cell. However, while this approach has yielded many significant discoveries, GFP and similar tags are so large that they may interfere with the labeled proteins’ natural functions.
A new approach based on cell-squeezing technology developed at MIT allows researchers to deliver fluorescent tags that are much less bulky, making this kind of protein imaging easier and more efficient.
In 2013, the MIT team demonstrated that squeezing cells makes it possible to deliver a variety of molecules, including proteins, DNA, carbon nanotubes, and quantum dots, into the cells without damaging them.
Researchers at Goethe University Frankfurt in Germany, working with their MIT colleagues, have now employed this approach to deliver relatively tiny fluorescent tags that can be targeted to specific proteins. Using regular confocal microscopes or super resolution microscopes, scientists can then track these proteins over time as they perform their normal functions.
“It really opens up the door to watching protein interactions in live cells,” says Armon Sharei, a former postdoc at MIT’s Koch Institute for Integrative Cancer Research. “Proteins are the building blocks of cells and control all their functions, so it’s exciting to be able to finally visualize them in a living cell, without genetic modifications.”
Sharei is an author of a paper describing the technique in Nature Communications. The paper’s lead author is Alina Kollmannsperger, a graduate student at Goethe University, and the senior authors are Ralph Wieneke and Robert Tampé of Goethe University. Robert Langer, the David H. Koch Institute Professor at MIT, and Klavs Jensen, the Warren K. Lewis Professor of Chemical Engineering at MIT, are also authors.
“We are very excited about this latest application for our cell squeezing approach and its implications for protein labeling,” says Langer, who is a member of MIT’s Koch Institute for Integrative Cancer Research.
Rapid delivery
In their 2013 study, the MIT team showed that squeezing cells through a constriction 30 to 80 percent smaller than the cells’ diameter caused tiny, temporary holes to appear in the cell membranes, allowing any large molecules in the surrounding fluid to enter. The holes reseal quickly and the cells suffer no long-term damage.
The researchers then began working with the Goethe University team to use this technique to label proteins with small fluorescent tags, which have previously been difficult to get into living cells. The Goethe team developed a tag called trisNTA that binds to any protein with a long string of histidine molecules (one of the 20 amino acids that form the building blocks of proteins).
For this study, the researchers first used genetic engineering to attach the histidine sequence to several different proteins, including one found in the nucleus and another involved in processing foreign molecules that have entered the cell. Then, the cells were pushed through a microfluidic channel at a rate of 1 million cells per second, which squeezed them sufficiently to allow the trisNTA tag in.
Until now, scientists have had to use protein tags, such as the bulky GFP, that can be genetically encoded in the cells’ DNA, or to study proteins in nonliving cells because the process of getting other fluorescent tags into the cells requires destroying the cell membrane.
“This study shows how microfluidic cell-squeezing together with specific chemical labeling can be exploited to hook various synthetic fluorophores to intracellular proteins with exquisite specificity. I foresee many applications for this approach and I have a very long list of probes that I would like to test immediately,” says Kai Johnsson, a professor of chemical sciences and engineering at the École Polytechnique Fédérale de Lausanne in Switzerland, who was not involved in the research.
With further work, including the development of new tags that target other proteins, this technique could help scientists learn much more about proteins’ functions inside living cells.
“Basically everything that happens in your cells is mediated by proteins,” Sharei says. “You can start to learn a lot about the basic biology of how a cell works, how it divides, and what makes the cancer cell a cancer cell, as far as what mechanisms go awry and what proteins are responsible for that.”
Normal cell behavior
The researchers believe that the cell squeezing technique should work with nearly any type of cell. So far, they have tried it successfully with more than 30 different types of mammalian cells.
An added benefit is that when cells undergo the squeezing procedure, they show no changes in the genes they express. In contrast, when a jolt of electricity is applied to cells to make them more permeable — a technique commonly used to deliver DNA and RNA — more than 7,000 genes are affected.
“It’s possible to assume that a squeezed cell is probably going to behave more or less normally, which is critical when you’re trying to study these kinds of processes,” Sharei says.
A company called SQZ Biotech, started by MIT researchers including Sharei, Langer, and Jensen, has licensed the cell squeezing technology and is now using it to engineer immune cells to improve their ability to attack cancer cells.